
| Name: LOC100133405 | Sequence: fasta or formatted (278aa) | NCBI GI: 239751469 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
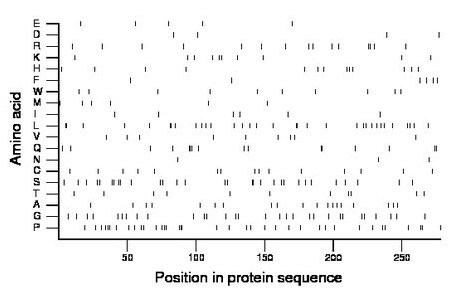
Amino acid Percentage Count Longest homopolymer A alanine 6.8 19 1 C cysteine 5.4 15 1 D aspartate 1.4 4 1 E glutamate 1.8 5 1 F phenylalanine 2.2 6 1 G glycine 10.8 30 1 H histidine 4.7 13 1 I isoleucine 2.2 6 1 K lysine 3.6 10 1 L leucine 11.9 33 2 M methionine 2.2 6 1 N asparagine 1.1 3 1 P proline 13.3 37 3 Q glutamine 5.8 16 3 R arginine 5.4 15 2 S serine 10.8 30 2 T threonine 4.0 11 1 V valine 3.2 9 1 W tryptophan 3.6 10 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein HES3 0.026 hairy and enhancer of split 3 WASF1 0.021 Wiskott-Aldrich syndrome protein family member 1 [Ho... WASF1 0.021 Wiskott-Aldrich syndrome protein family member 1 [Ho... WASF1 0.021 Wiskott-Aldrich syndrome protein family member 1 [Ho... WASF1 0.021 Wiskott-Aldrich syndrome protein family member 1 [Hom... HCN4 0.017 hyperpolarization activated cyclic nucleotide-gated p... DIAPH1 0.017 diaphanous 1 isoform 2 DIAPH1 0.017 diaphanous 1 isoform 1 SPEG 0.017 SPEG complex locus SMARCC1 0.015 SWI/SNF-related matrix-associated actin-dependent r... LOC100127987 0.014 PREDICTED: hypothetical protein LOC100127987 LOC100127987 0.014 PREDICTED: hypothetical protein LOC100127987 LOC100127987 0.014 PREDICTED: hypothetical protein LOC100127987 TAF4 0.014 TBP-associated factor 4 VPS37B 0.012 vacuolar protein sorting 37B MYO15A 0.012 myosin XV TBKBP1 0.012 TBK1 binding protein 1 WNK2 0.012 WNK lysine deficient protein kinase 2 ALPK3 0.012 alpha-kinase 3 RGL4 0.012 ral guanine nucleotide dissociation stimulator-like ... LOC100292287 0.012 PREDICTED: hypothetical protein LOC100289576 0.012 PREDICTED: hypothetical protein LOC100289576 0.012 PREDICTED: hypothetical protein XP_002343622 TET3 0.012 tet oncogene family member 3 BBC3 0.012 BCL2 binding component 3 isoform 1 FAM134B 0.012 hypothetical protein LOC54463 isoform 1 SNAP91 0.010 synaptosomal-associated protein, 91kDa homolog LOC100288699 0.010 PREDICTED: hypothetical protein XP_002343846 LOC653550 0.010 PREDICTED: similar to TP53 target 3 isoform 3 [Homo...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
