
| Name: LOC100290663 | Sequence: fasta or formatted (72aa) | NCBI GI: 239751148 | |
|
Description: PREDICTED: hypothetical protein XP_002347729
| Not currently referenced in the text | ||
|
Composition:
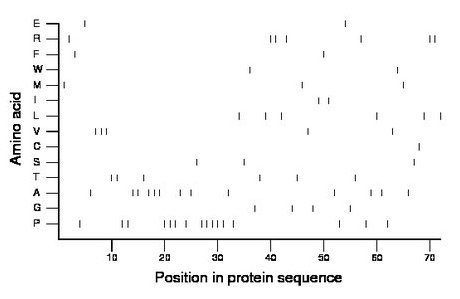
Amino acid Percentage Count Longest homopolymer A alanine 18.1 13 3 C cysteine 1.4 1 1 D aspartate 0.0 0 0 E glutamate 2.8 2 1 F phenylalanine 2.8 2 1 G glycine 5.6 4 1 H histidine 0.0 0 0 I isoleucine 2.8 2 1 K lysine 0.0 0 0 L leucine 8.3 6 1 M methionine 4.2 3 1 N asparagine 0.0 0 0 P proline 22.2 16 5 Q glutamine 0.0 0 0 R arginine 9.7 7 2 S serine 4.2 3 1 T threonine 8.3 6 2 V valine 6.9 5 3 W tryptophan 2.8 2 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002347729 SAMD1 0.142 sterile alpha motif domain containing 1 SEZ6L2 0.118 seizure related 6 homolog (mouse)-like 2 isoform 4 ... SEZ6L2 0.118 seizure related 6 homolog (mouse)-like 2 isoform 2 ... TAF4 0.110 TBP-associated factor 4 USP51 0.110 ubiquitin specific protease 51 HOXD8 0.094 homeobox D8 KIAA1949 0.094 phostensin KIAA1949 0.094 phostensin KDM6B 0.094 lysine (K)-specific demethylase 6B DGKD 0.094 diacylglycerol kinase, delta 130kDa isoform 2 FOXK1 0.094 forkhead box K1 DIAPH1 0.094 diaphanous 1 isoform 2 DIAPH1 0.094 diaphanous 1 isoform 1 BAT3 0.094 HLA-B associated transcript-3 isoform b BAT3 0.094 HLA-B associated transcript-3 isoform b BAT3 0.094 HLA-B associated transcript-3 isoform b BAT3 0.094 HLA-B associated transcript-3 isoform a SFRS15 0.094 splicing factor, arginine/serine-rich 15 isoform 3 ... SFRS15 0.094 splicing factor, arginine/serine-rich 15 isoform 2 ... SFRS15 0.094 splicing factor, arginine/serine-rich 15 isoform 1 [... MAP3K4 0.094 mitogen-activated protein kinase kinase kinase 4 iso... MAP3K4 0.094 mitogen-activated protein kinase kinase kinase 4 iso... RANBP9 0.087 RAN binding protein 9 LOC645529 0.087 PREDICTED: similar to Putative acrosin-like proteas... STK39 0.087 serine threonine kinase 39 (STE20/SPS1 homolog, yea... SHANK3 0.087 SH3 and multiple ankyrin repeat domains 3 INHBB 0.087 inhibin beta B subunit preproprotein PRR12 0.087 proline rich 12 TRRAP 0.079 transformation/transcription domain-associated protei...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
