
| Name: CMPK2 | Sequence: fasta or formatted (449aa) | NCBI GI: 117606370 | |
|
Description: UMP-CMP kinase 2
|
Referenced in:
| ||
|
Composition:
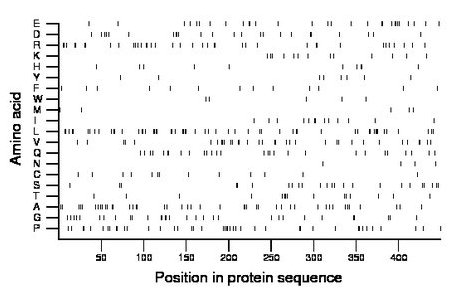
Amino acid Percentage Count Longest homopolymer A alanine 9.8 44 2 C cysteine 2.9 13 1 D aspartate 5.3 24 2 E glutamate 6.5 29 2 F phenylalanine 2.9 13 1 G glycine 6.9 31 2 H histidine 2.2 10 2 I isoleucine 2.9 13 2 K lysine 2.9 13 1 L leucine 12.5 56 3 M methionine 1.1 5 1 N asparagine 0.9 4 1 P proline 9.4 42 2 Q glutamine 6.2 28 2 R arginine 8.5 38 2 S serine 5.3 24 2 T threonine 3.3 15 2 V valine 7.6 34 2 W tryptophan 1.1 5 1 Y tyrosine 1.8 8 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 UMP-CMP kinase 2 TK2 0.015 thymidine kinase 2, mitochondrial SLC9A1 0.009 solute carrier family 9, isoform A1 LOC100292117 0.009 PREDICTED: hypothetical protein XP_002344623 LOC100288224 0.009 PREDICTED: hypothetical protein LOC100288224 0.009 PREDICTED: hypothetical protein XP_002343139 GGA1 0.008 golgi associated, gamma adaptin ear containing, ARF b... THSD7B 0.008 thrombospondin, type I, domain containing 7B LOC645781 0.008 PREDICTED: similar to mCG141871 LOC645781 0.008 PREDICTED: similar to mCG141871 LOC645781 0.008 PREDICTED: similar to mCG141871 BAT2 0.007 HLA-B associated transcript-2 ALS2CR4 0.007 amyotrophic lateral sclerosis 2 (juvenile) chromoso... ALS2CR4 0.007 amyotrophic lateral sclerosis 2 (juvenile) chromoso... MGAT1 0.007 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... MGAT1 0.007 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... MGAT1 0.007 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... MGAT1 0.007 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... MGAT1 0.007 mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acety... SSPO 0.007 SCO-spondin RPTOR 0.006 raptor isoform 1 RPTOR 0.006 raptor isoform 2 GGA1 0.006 golgi associated, gamma adaptin ear containing, ARF ... C2orf21 0.006 chromosome 2 open reading frame 21 isoform 1 LOC401180 0.005 PREDICTED: similar to hCG1745223 LOC401180 0.005 PREDICTED: similar to hCG1745223 LOC401180 0.005 PREDICTED: similar to hCG1745223 HEPACAM 0.005 hepatocyte cell adhesion molecule PRR18 0.005 proline rich region 18 RICS 0.005 Rho GTPase-activating protein isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
