
| Name: LOC100290011 | Sequence: fasta or formatted (242aa) | NCBI GI: 239749305 | |
|
Description: PREDICTED: hypothetical protein XP_002346991
| Not currently referenced in the text | ||
|
Composition:
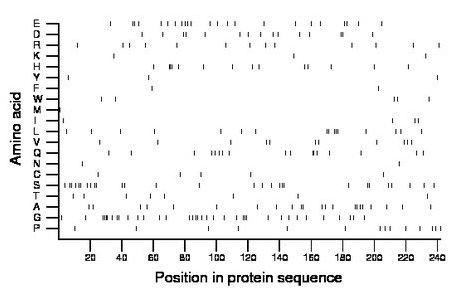
Amino acid Percentage Count Longest homopolymer A alanine 9.1 22 2 C cysteine 2.1 5 1 D aspartate 6.2 15 2 E glutamate 9.5 23 2 F phenylalanine 0.8 2 1 G glycine 14.9 36 4 H histidine 5.8 14 3 I isoleucine 1.7 4 1 K lysine 1.2 3 1 L leucine 6.6 16 3 M methionine 0.4 1 1 N asparagine 0.8 2 1 P proline 5.8 14 1 Q glutamine 7.0 17 2 R arginine 5.4 13 1 S serine 11.6 28 2 T threonine 3.3 8 1 V valine 4.5 11 1 W tryptophan 2.1 5 1 Y tyrosine 1.2 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346991 LOC100292821 1.000 PREDICTED: hypothetical protein LOC100287222 1.000 PREDICTED: hypothetical protein XP_002342839 LOC100287222 1.000 PREDICTED: hypothetical protein RPGR 0.050 retinitis pigmentosa GTPase regulator isoform C [Hom... RP1L1 0.043 retinitis pigmentosa 1-like 1 FLG 0.043 filaggrin SLC24A1 0.039 solute carrier family 24 (sodium/potassium/calcium ex... AEBP2 0.037 AE binding protein 2 isoform b AEBP2 0.037 AE binding protein 2 isoform a TPR 0.033 nuclear pore complex-associated protein TPR PPP1R10 0.031 protein phosphatase 1, regulatory subunit 10 LEO1 0.031 Leo1, Paf1/RNA polymerase II complex component, homo... HNRNPU 0.031 heterogeneous nuclear ribonucleoprotein U isoform b ... HNRNPU 0.031 heterogeneous nuclear ribonucleoprotein U isoform a ... CASC3 0.031 metastatic lymph node 51 SPARCL1 0.029 SPARC-like 1 SPARCL1 0.029 SPARC-like 1 HRNR 0.027 hornerin IWS1 0.023 IWS1 homolog LOC100292287 0.021 PREDICTED: hypothetical protein LOC100289576 0.021 PREDICTED: hypothetical protein LOC100289576 0.021 PREDICTED: hypothetical protein XP_002343622 NEFM 0.021 neurofilament, medium polypeptide 150kDa isoform 1 ... NEFM 0.021 neurofilament, medium polypeptide 150kDa isoform 2 ... CLIC6 0.021 chloride intracellular channel 6 CHGB 0.021 chromogranin B precursor ABLIM2 0.021 actin binding LIM protein family, member 2 isoform ... ABLIM2 0.021 actin binding LIM protein family, member 2 isoform ... RBM15B 0.019 RNA binding motif protein 15BHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
