
| Name: LOC100290658 | Sequence: fasta or formatted (76aa) | NCBI GI: 239749201 | |
|
Description: PREDICTED: hypothetical protein XP_002346947
| Not currently referenced in the text | ||
|
Composition:
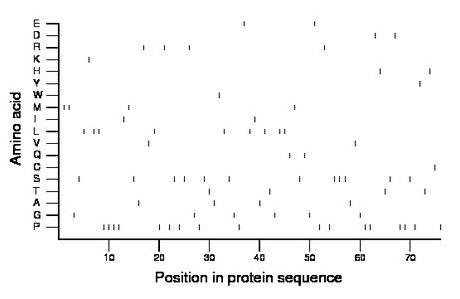
Amino acid Percentage Count Longest homopolymer A alanine 5.3 4 1 C cysteine 1.3 1 1 D aspartate 2.6 2 1 E glutamate 2.6 2 1 F phenylalanine 0.0 0 0 G glycine 7.9 6 1 H histidine 2.6 2 1 I isoleucine 2.6 2 1 K lysine 1.3 1 1 L leucine 11.8 9 2 M methionine 5.3 4 2 N asparagine 0.0 0 0 P proline 22.4 17 4 Q glutamine 2.6 2 1 R arginine 5.3 4 1 S serine 15.8 12 3 T threonine 5.3 4 1 V valine 2.6 2 1 W tryptophan 1.3 1 1 Y tyrosine 1.3 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346947 LOC100293864 1.000 PREDICTED: hypothetical protein LOC100289367 1.000 PREDICTED: hypothetical protein XP_002342784 LOC100289367 1.000 PREDICTED: similar to FLJ00248 protein SCARF2 0.073 scavenger receptor class F, member 2 isoform 1 [Homo... SCARF2 0.073 scavenger receptor class F, member 2 isoform 2 [Homo... LRP10 0.066 low density lipoprotein receptor-related protein 10 ... WASL 0.058 Wiskott-Aldrich syndrome gene-like protein SETD1B 0.051 SET domain containing 1B RANBP9 0.051 RAN binding protein 9 LOC100132891 0.051 PREDICTED: hypothetical protein LOC100132891 0.051 PREDICTED: hypothetical protein LOC100132891 0.051 PREDICTED: hypothetical protein KIAA0913 0.051 hypothetical protein LOC23053 SRCAP 0.051 Snf2-related CBP activator protein FLJ22184 0.044 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.044 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.044 PREDICTED: hypothetical protein LOC80164 FBLIM1 0.044 filamin-binding LIM protein-1 isoform b FBLIM1 0.044 filamin-binding LIM protein-1 isoform a PLK3 0.044 polo-like kinase 3 DMRT3 0.044 doublesex and mab-3 related transcription factor 3 [... CIC 0.044 capicua homolog FCHSD1 0.044 FCH and double SH3 domains 1 MBD6 0.044 methyl-CpG binding domain protein 6 FMN1 0.044 formin 1 ZNF358 0.044 zinc finger protein 358 SMARCA4 0.036 SWI/SNF-related matrix-associated actin-dependent r... SMARCA4 0.036 SWI/SNF-related matrix-associated actin-dependent r... SMARCA4 0.036 SWI/SNF-related matrix-associated actin-dependent r...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
