
| Name: LOC100289753 | Sequence: fasta or formatted (80aa) | NCBI GI: 239747543 | |
|
Description: PREDICTED: hypothetical protein XP_002347646
| Not currently referenced in the text | ||
|
Composition:
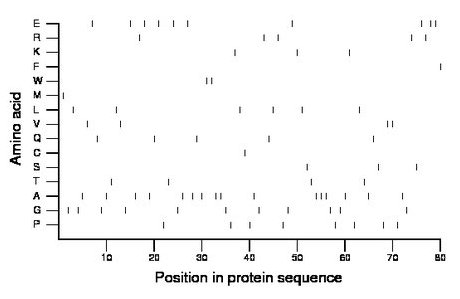
Amino acid Percentage Count Longest homopolymer A alanine 20.0 16 3 C cysteine 1.2 1 1 D aspartate 0.0 0 0 E glutamate 12.5 10 2 F phenylalanine 1.2 1 1 G glycine 13.8 11 1 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 3.8 3 1 L leucine 7.5 6 1 M methionine 1.2 1 1 N asparagine 0.0 0 0 P proline 10.0 8 1 Q glutamine 6.2 5 1 R arginine 6.2 5 1 S serine 3.8 3 1 T threonine 5.0 4 1 V valine 5.0 4 2 W tryptophan 2.5 2 2 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002347646 LOC100293292 0.597 PREDICTED: hypothetical protein LOC100287612 0.597 PREDICTED: hypothetical protein XP_002342706 LOC100131199 0.065 PREDICTED: hypothetical protein C17orf96 0.058 hypothetical protein LOC100170841 BSN 0.050 bassoon protein LOC100132249 0.036 PREDICTED: hypothetical protein DLST 0.036 dihydrolipoamide S-succinyltransferase (E2 component... RP1L1 0.036 retinitis pigmentosa 1-like 1 MUPCDH 0.036 mucin and cadherin-like isoform 1 PNCK 0.029 pregnancy upregulated non-ubiquitously expressed Ca... TAF4 0.029 TBP-associated factor 4 LOC100288476 0.022 PREDICTED: hypothetical protein XP_002343815 LOC643395 0.022 PREDICTED: hypothetical protein MCTP1 0.022 multiple C2 domains, transmembrane 1 isoform L [Hom... FGF2 0.022 fibroblast growth factor 2 LOC100293144 0.022 PREDICTED: hypothetical protein LOC100288857 0.022 PREDICTED: hypothetical protein LOC100288857 0.022 PREDICTED: hypothetical protein XP_002343308 CSPG5 0.022 chondroitin sulfate proteoglycan 5 (neuroglycan C) ... ZIC2 0.022 zinc finger protein of the cerebellum 2 FLJ10404 0.022 hypothetical protein LOC54540 LOC390595 0.022 PREDICTED: similar to ubiquitin-associated protein ... LOC100293898 0.022 PREDICTED: hypothetical protein LOC390595 0.022 PREDICTED: similar to ubiquitin-associated protein ... LOC100290946 0.022 PREDICTED: hypothetical protein XP_002347121 LOC390595 0.022 PREDICTED: hypothetical protein LOC100288840 0.022 PREDICTED: hypothetical protein XP_002344246 SHANK3 0.022 SH3 and multiple ankyrin repeat domains 3 LBXCOR1 0.022 LBXCOR1 homologHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
