
| Name: LOC100294140 | Sequence: fasta or formatted (125aa) | NCBI GI: 239740677 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
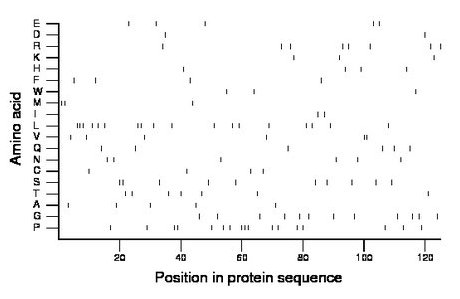
Amino acid Percentage Count Longest homopolymer A alanine 4.0 5 1 C cysteine 3.2 4 1 D aspartate 1.6 2 1 E glutamate 4.0 5 1 F phenylalanine 3.2 4 1 G glycine 9.6 12 1 H histidine 3.2 4 1 I isoleucine 1.6 2 1 K lysine 2.4 3 1 L leucine 14.4 18 3 M methionine 2.4 3 2 N asparagine 4.8 6 1 P proline 13.6 17 3 Q glutamine 4.8 6 1 R arginine 6.4 8 1 S serine 8.0 10 2 T threonine 5.6 7 1 V valine 4.8 6 2 W tryptophan 2.4 3 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100294242 1.000 PREDICTED: hypothetical protein LOC100294129 1.000 PREDICTED: hypothetical protein LOC100293661 0.996 PREDICTED: hypothetical protein LOC100289810 0.996 PREDICTED: hypothetical protein XP_002346828 LOC100287329 0.996 PREDICTED: hypothetical protein XP_002342661 LOC100294294 0.996 PREDICTED: hypothetical protein XP_002344096 LOC100294254 0.926 PREDICTED: hypothetical protein XP_002344068 CDAN1 0.029 codanin 1 CRLS1 0.025 cardiolipin synthase 1 isoform 1 TICAM1 0.020 toll-like receptor adaptor molecule 1 EGR1 0.016 early growth response 1 NUP214 0.016 nucleoporin 214kDa KRT38 0.016 keratin 38 PLXNB1 0.016 plexin B1 PLXNB1 0.016 plexin B1 ADAMTS7 0.016 ADAM metallopeptidase with thrombospondin type 1 mot... CPSF6 0.016 cleavage and polyadenylation specific factor 6, 68 ... PRR18 0.016 proline rich region 18 MED9 0.016 mediator complex subunit 9 PLEKHA7 0.012 pleckstrin homology domain containing, family A mem... LOC100291797 0.012 PREDICTED: hypothetical protein XP_002345651 LOC100291443 0.012 PREDICTED: hypothetical protein XP_002346504 LOC100289394 0.012 PREDICTED: hypothetical protein XP_002342316 BCL2L13 0.012 BCL2-like 13 (apoptosis facilitator) LEF1 0.012 lymphoid enhancer-binding factor 1 isoform 1 LOC100292629 0.012 PREDICTED: hypothetical protein LOC100128602 0.012 PREDICTED: hypothetical protein FLJ22184 0.012 PREDICTED: hypothetical protein FLJ22184 LOC100289960 0.012 PREDICTED: hypothetical protein XP_002346853Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
