
| Name: CRLS1 | Sequence: fasta or formatted (301aa) | NCBI GI: 10092647 | |
|
Description: cardiolipin synthase 1 isoform 1
|
Referenced in:
| ||
Other entries for this name:
alt prot [202aa] cardiolipin synthase 1 isoform 2 | |||
|
Composition:
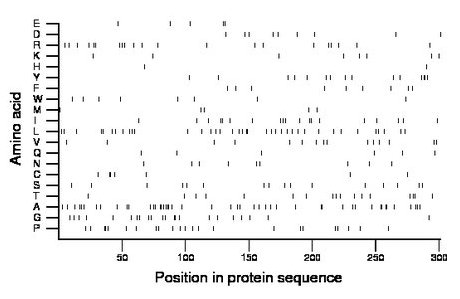
Amino acid Percentage Count Longest homopolymer A alanine 15.6 47 3 C cysteine 2.3 7 2 D aspartate 3.0 9 1 E glutamate 1.7 5 2 F phenylalanine 3.0 9 1 G glycine 7.3 22 2 H histidine 0.7 2 1 I isoleucine 6.3 19 2 K lysine 2.7 8 1 L leucine 12.6 38 2 M methionine 1.7 5 1 N asparagine 3.0 9 1 P proline 7.6 23 2 Q glutamine 2.0 6 1 R arginine 7.0 21 2 S serine 5.3 16 1 T threonine 5.6 17 2 V valine 5.0 15 1 W tryptophan 2.7 8 1 Y tyrosine 5.0 15 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 cardiolipin synthase 1 isoform 1 CRLS1 0.630 cardiolipin synthase 1 isoform 2 ITPKC 0.017 inositol 1,4,5-trisphosphate 3-kinase C CDIPT 0.015 CDP-diacylglycerol-inositol 3-phosphatidyltransferase... SCAF1 0.015 SR-related CTD-associated factor 1 VAX1 0.014 ventral anterior homeobox 1 isoform a ESYT2 0.014 family with sequence similarity 62 (C2 domain contai... LOC100291634 0.014 PREDICTED: hypothetical protein XP_002346062 LOC100291096 0.014 PREDICTED: hypothetical protein XP_002346952 LOC100287232 0.014 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.014 PREDICTED: hypothetical protein HECA 0.014 headcase LOC100128572 0.012 PREDICTED: hypothetical protein LOC100291220 0.012 PREDICTED: hypothetical protein XP_002347633 LOC100128572 0.012 PREDICTED: hypothetical protein LOC100287224 0.012 PREDICTED: hypothetical protein XP_002343483 LOC100128572 0.012 PREDICTED: hypothetical protein C17orf65 0.012 hypothetical protein LOC339201 LOC100292958 0.012 PREDICTED: hypothetical protein LOC100293661 0.012 PREDICTED: hypothetical protein LOC100288311 0.012 PREDICTED: hypothetical protein LOC100289810 0.012 PREDICTED: hypothetical protein XP_002346828 LOC100288311 0.012 PREDICTED: hypothetical protein XP_002342972 LOC100287329 0.012 PREDICTED: hypothetical protein XP_002342661 LOC100294294 0.012 PREDICTED: hypothetical protein XP_002344096 IER5L 0.012 immediate early response 5-like LOC100290265 0.012 PREDICTED: hypothetical protein XP_002346787 LOC727804 0.012 PREDICTED: hypothetical protein TLX1 0.012 T-cell leukemia homeobox 1 SMARCD2 0.012 SWI/SNF-related matrix-associated actin-dependent r...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
