
| Name: GPSM3 | Sequence: fasta or formatted (160aa) | NCBI GI: 11545817 | |
|
Description: G-protein signaling modulator 3 (AGS3-like, C. elegans)
|
Referenced in:
| ||
|
Composition:
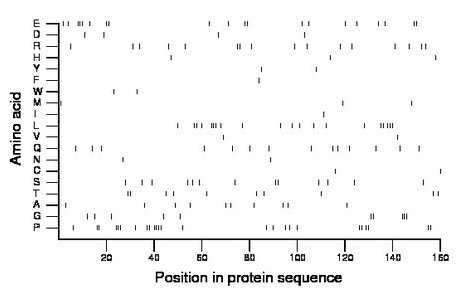
Amino acid Percentage Count Longest homopolymer A alanine 6.2 10 1 C cysteine 1.2 2 1 D aspartate 2.5 4 1 E glutamate 12.5 20 3 F phenylalanine 0.6 1 1 G glycine 6.2 10 3 H histidine 1.9 3 1 I isoleucine 0.6 1 1 K lysine 0.0 0 0 L leucine 12.5 20 3 M methionine 1.9 3 1 N asparagine 1.2 2 1 P proline 15.6 25 4 Q glutamine 8.8 14 1 R arginine 10.0 16 2 S serine 8.1 13 2 T threonine 6.2 10 2 V valine 1.2 2 1 W tryptophan 1.2 2 1 Y tyrosine 1.2 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 G-protein signaling modulator 3 (AGS3-like, C. elega... GPSM1 0.078 G-protein signaling modulator 1 (AGS3-like, C. eleg... GPSM1 0.078 G-protein signaling modulator 1 (AGS3-like, C. eleg... GPSM2 0.065 LGN protein PWWP2A 0.052 PWWP domain containing 2A isoform b PWWP2A 0.052 PWWP domain containing 2A isoform a CIC 0.045 capicua homolog SFI1 0.039 spindle assembly associated Sfi1 homolog isoform a [... SFI1 0.039 spindle assembly associated Sfi1 homolog isoform b [... FLJ20021 0.039 PREDICTED: hypothetical protein FLJ20021 0.039 PREDICTED: hypothetical protein FLJ20021 0.039 PREDICTED: hypothetical protein CCDC64B 0.036 coiled-coil domain containing 64B PRR12 0.036 proline rich 12 LRRC37B 0.036 leucine rich repeat containing 37B WAS 0.032 Wiskott-Aldrich syndrome protein SOCS7 0.032 suppressor of cytokine signaling 7 FAM148B 0.032 nuclear localized factor 2 IRS2 0.032 insulin receptor substrate 2 RBBP6 0.032 retinoblastoma-binding protein 6 isoform 2 HNRNPUL2 0.032 heterogeneous nuclear ribonucleoprotein U-like 2 [H... KIF18B 0.032 kinesin family member 18B C6orf132 0.029 PREDICTED: chromosome 6 open reading frame 132 [Hom... C6orf132 0.029 PREDICTED: chromosome 6 open reading frame 132 [Hom... C6orf132 0.029 PREDICTED: hypothetical protein LOC647024 SCRIB 0.029 scribble isoform b SCRIB 0.029 scribble isoform a SYNJ1 0.029 synaptojanin 1 isoform d PLEKHO1 0.029 pleckstrin homology domain containing, family O memb... BCL6B 0.029 B-cell CLL/lymphoma 6, member B (zinc finger protein...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
