
| Name: LOC440498 | Sequence: fasta or formatted (74aa) | NCBI GI: 209969760 | |
|
Description: heat shock factor binding protein 1-like
| Not currently referenced in the text | ||
|
Composition:
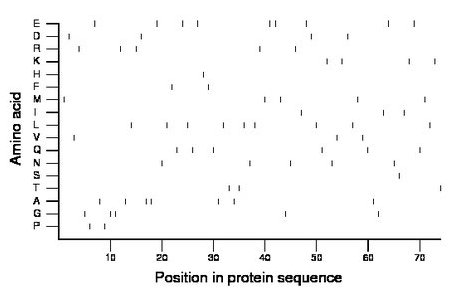
Amino acid Percentage Count Longest homopolymer A alanine 9.5 7 2 C cysteine 0.0 0 0 D aspartate 5.4 4 1 E glutamate 12.2 9 2 F phenylalanine 2.7 2 1 G glycine 6.8 5 2 H histidine 1.4 1 1 I isoleucine 4.1 3 1 K lysine 5.4 4 1 L leucine 12.2 9 1 M methionine 6.8 5 1 N asparagine 6.8 5 1 P proline 2.7 2 1 Q glutamine 8.1 6 1 R arginine 6.8 5 1 S serine 1.4 1 1 T threonine 4.1 3 1 V valine 4.1 3 1 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 heat shock factor binding protein 1-like HSBP1 0.213 heat shock factor binding protein 1 LOC100291861 0.172 PREDICTED: similar to heat shock factor binding pro... LOC100290361 0.172 PREDICTED: hypothetical protein XP_002346659 LOC100288483 0.172 PREDICTED: hypothetical protein XP_002342506 APOE 0.066 apolipoprotein E precursor TH1L 0.066 TH1-like protein FLJ20674 0.041 hypothetical protein FLJ20674 AKAP9 0.041 A-kinase anchor protein 9 isoform 2 AKAP9 0.041 A-kinase anchor protein 9 isoform 3 KIF27 0.033 kinesin family member 27 ANKRD26 0.033 ankyrin repeat domain 26 CGN 0.025 cingulin ATP8A1 0.025 ATPase, aminophospholipid transporter (APLT), class... ATP8A1 0.025 ATPase, aminophospholipid transporter (APLT), class ... HOOK3 0.025 golgi-associated microtubule-binding protein HOOK3 [... FAM9A 0.025 family with sequence similarity 9, member A C6orf163 0.016 PREDICTED: chromosome 6 open reading frame 163 [Hom... C6orf163 0.016 PREDICTED: chromosome 6 open reading frame 163 [Hom... C6orf163 0.016 PREDICTED: hypothetical protein LOC206412 APPL1 0.016 adaptor protein, phosphotyrosine interaction, PH doma... LOC342346 0.016 hypothetical protein LOC342346 TNIP1 0.016 TNFAIP3 interacting protein 1 RNF20 0.016 ring finger protein 20 PPFIA3 0.016 PTPRF interacting protein alpha 3 GCC2 0.016 GRIP and coiled-coil domain-containing 2 CEP290 0.016 centrosomal protein 290kDa SYNE1 0.016 spectrin repeat containing, nuclear envelope 1 isofo... SYNE1 0.016 spectrin repeat containing, nuclear envelope 1 isof... CENPF 0.016 centromere protein FHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
