
| Name: LOC100127894 | Sequence: fasta or formatted (331aa) | NCBI GI: 169173235 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
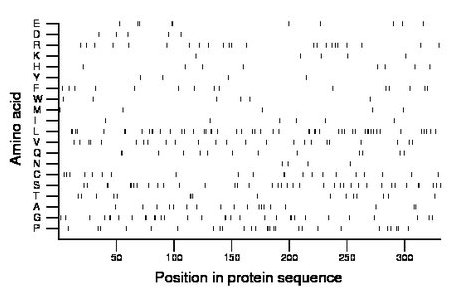
Amino acid Percentage Count Longest homopolymer A alanine 5.4 18 1 C cysteine 6.3 21 2 D aspartate 1.5 5 1 E glutamate 3.0 10 2 F phenylalanine 4.5 15 1 G glycine 9.7 32 2 H histidine 2.4 8 1 I isoleucine 1.8 6 1 K lysine 1.5 5 1 L leucine 15.4 51 2 M methionine 1.5 5 1 N asparagine 1.2 4 1 P proline 8.2 27 3 Q glutamine 3.9 13 2 R arginine 6.9 23 1 S serine 11.5 38 2 T threonine 4.8 16 3 V valine 7.3 24 1 W tryptophan 1.8 6 1 Y tyrosine 1.2 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC730413 0.092 PREDICTED: hypothetical protein LOC730413 0.092 PREDICTED: hypothetical protein LOC100293459 0.065 PREDICTED: similar to hCG1984907 LOC100293174 0.024 PREDICTED: similar to transmembrane phosphoinositid... ZFHX3 0.014 AT-binding transcription factor 1 CECR6 0.012 cat eye syndrome chromosome region, candidate 6 iso... CECR6 0.012 cat eye syndrome chromosome region, candidate 6 isof... NSUN7 0.011 NOL1/NOP2/Sun domain family, member 7 BAHD1 0.011 bromo adjacent homology domain containing 1 TMEM158 0.011 Ras-induced senescence 1 ESPN 0.009 espin LOC100289886 0.009 PREDICTED: hypothetical protein XP_002348008 LOC100287120 0.009 PREDICTED: hypothetical protein XP_002343728 LPPR4 0.009 plasticity related gene 1 LOC100288439 0.008 PREDICTED: hypothetical protein LOC100288439 0.008 PREDICTED: hypothetical protein XP_002343141 PLEKHA5 0.008 pleckstrin homology domain containing, family A memb... ZNF728 0.006 PREDICTED: zinc finger protein 728 ZNF728 0.006 PREDICTED: zinc finger protein 728 ZNF728 0.006 PREDICTED: zinc finger protein 728 LOC401321 0.006 PREDICTED: hypothetical protein LOC401321 0.006 PREDICTED: hypothetical protein LOC401321 0.006 PREDICTED: hypothetical protein LOC401321 0.006 PREDICTED: hypothetical protein CPSF7 0.006 pre-mRNA cleavage factor I, 59 kDa subunit isoform ... CPSF7 0.006 pre-mRNA cleavage factor I, 59 kDa subunit isoform ... CPSF7 0.006 pre-mRNA cleavage factor I, 59 kDa subunit isoform ... POU2F3 0.005 POU transcription factor RHBDD2 0.005 rhomboid domain containing 2 isoform bHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
