
| Name: DCD | Sequence: fasta or formatted (110aa) | NCBI GI: 16751921 | |
|
Description: dermcidin preproprotein
|
Referenced in: Additional Host Defense Functions
| ||
|
Composition:
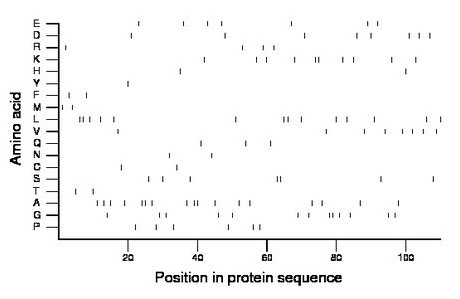
Amino acid Percentage Count Longest homopolymer A alanine 15.5 17 2 C cysteine 1.8 2 1 D aspartate 7.3 8 1 E glutamate 6.4 7 1 F phenylalanine 1.8 2 1 G glycine 11.8 13 2 H histidine 1.8 2 1 I isoleucine 0.0 0 0 K lysine 9.1 10 2 L leucine 12.7 14 2 M methionine 1.8 2 1 N asparagine 1.8 2 1 P proline 5.5 6 1 Q glutamine 2.7 3 1 R arginine 3.6 4 1 S serine 6.4 7 2 T threonine 1.8 2 1 V valine 7.3 8 1 W tryptophan 0.0 0 0 Y tyrosine 0.9 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 dermcidin preproprotein LACRT 0.057 lacritin precursor C2orf55 0.036 hypothetical protein LOC343990 C9orf140 0.026 tumor specificity and mitosis phase-dependent expre... TCEB3CL 0.026 transcription elongation factor B polypeptide 3C-li... TCEB3C 0.026 transcription elongation factor B polypeptide 3C [H... PHGDH 0.021 phosphoglycerate dehydrogenase KLF16 0.021 BTE-binding protein 4 LOC100130174 0.021 PREDICTED: hypothetical protein LOC100130174 0.021 PREDICTED: hypothetical protein KIAA1614 0.021 hypothetical protein LOC57710 INPP5D 0.021 SH2 containing inositol phosphatase isoform b INPP5D 0.021 SH2 containing inositol phosphatase isoform a OTUD1 0.015 OTU domain containing 1 SFRS18 0.015 splicing factor, arginine/serine-rich 130 SFRS18 0.015 splicing factor, arginine/serine-rich 130 MARCKS 0.015 myristoylated alanine-rich protein kinase C substra... SPG20 0.015 spartin SPG20 0.015 spartin SPG20 0.015 spartin SPG20 0.015 spartin STK38L 0.015 serine/threonine kinase 38 like PNMAL2 0.015 PNMA-like 2 LOC100128960 0.015 PREDICTED: similar to paraneoplastic antigen MA2 [H... LOC100128960 0.015 PREDICTED: similar to Putative paraneoplastic antig... TCOF1 0.015 Treacher Collins-Franceschetti syndrome 1 isoform f... TCOF1 0.015 Treacher Collins-Franceschetti syndrome 1 isoform e... TCOF1 0.015 Treacher Collins-Franceschetti syndrome 1 isoform d... TCOF1 0.015 Treacher Collins-Franceschetti syndrome 1 isoform b ... TCOF1 0.015 Treacher Collins-Franceschetti syndrome 1 isoform a ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
