
| Name: KIAA0020 | Sequence: fasta or formatted (648aa) | NCBI GI: 109948283 | |
|
Description: KIAA0020 protein
| Not currently referenced in the text | ||
|
Composition:
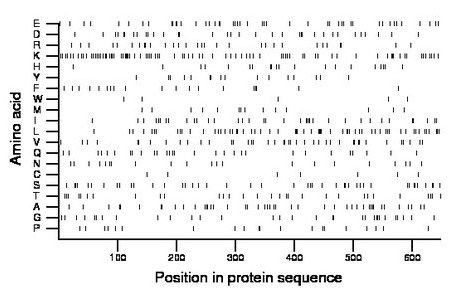
Amino acid Percentage Count Longest homopolymer A alanine 7.1 46 2 C cysteine 1.1 7 1 D aspartate 5.2 34 2 E glutamate 7.1 46 2 F phenylalanine 2.9 19 2 G glycine 4.9 32 2 H histidine 3.1 20 1 I isoleucine 6.0 39 3 K lysine 13.6 88 5 L leucine 9.6 62 2 M methionine 2.3 15 1 N asparagine 3.2 21 1 P proline 3.1 20 2 Q glutamine 4.6 30 2 R arginine 4.8 31 3 S serine 6.9 45 2 T threonine 4.6 30 1 V valine 6.5 42 2 W tryptophan 0.8 5 1 Y tyrosine 2.5 16 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 KIAA0020 protein PUM1 0.038 pumilio 1 isoform 2 PUM1 0.036 pumilio 1 isoform 1 PUM2 0.035 pumilio homolog 2 LOC100133599 0.014 PREDICTED: hypothetical protein FAM133B 0.012 hypothetical protein LOC257415 isoform 2 FAM133B 0.012 hypothetical protein LOC257415 isoform 1 CYLC2 0.012 cylicin 2 ASPH 0.012 aspartate beta-hydroxylase isoform d ASPH 0.012 aspartate beta-hydroxylase isoform e ANKRD12 0.012 ankyrin repeat domain 12 isoform 2 ANKRD12 0.012 ankyrin repeat domain 12 isoform 1 TRAF3IP1 0.011 TNF receptor-associated factor 3 interacting protei... TRAF3IP1 0.011 TNF receptor-associated factor 3 interacting protein... SFRS11 0.010 splicing factor, arginine/serine-rich 11 FAM133A 0.010 hypothetical protein LOC286499 RNF20 0.010 ring finger protein 20 LOC731605 0.010 PREDICTED: similar to BCL2-associated transcription... LOC731605 0.010 PREDICTED: similar to BCL2-associated transcription... LOC100133599 0.009 PREDICTED: hypothetical protein BCLAF1 0.009 BCL2-associated transcription factor 1 isoform 3 [H... BCLAF1 0.009 BCL2-associated transcription factor 1 isoform 2 [H... BCLAF1 0.009 BCL2-associated transcription factor 1 isoform 1 [Hom... NKTR 0.009 natural killer-tumor recognition sequence PCF11 0.009 pre-mRNA cleavage complex II protein Pcf11 HTATSF1 0.008 HIV-1 Tat specific factor 1 HTATSF1 0.008 HIV-1 Tat specific factor 1 MYO10 0.008 myosin X SMC1A 0.008 structural maintenance of chromosomes 1A BAZ1B 0.008 bromodomain adjacent to zinc finger domain, 1B [Homo...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
