
| Name: SEPHS2 | Sequence: fasta or formatted (448aa) | NCBI GI: 15011844 | |
|
Description: selenophosphate synthetase 2
|
Referenced in:
| ||
|
Composition:
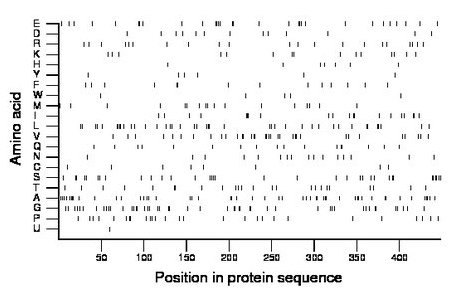
Amino acid Percentage Count Longest homopolymer A alanine 11.2 50 3 C cysteine 1.8 8 1 D aspartate 2.9 13 1 E glutamate 6.7 30 3 F phenylalanine 3.1 14 2 G glycine 10.7 48 2 H histidine 1.6 7 1 I isoleucine 4.5 20 3 K lysine 3.3 15 1 L leucine 9.2 41 2 M methionine 3.8 17 2 N asparagine 3.6 16 1 P proline 6.0 27 2 Q glutamine 3.8 17 2 R arginine 4.7 21 1 S serine 7.6 34 2 T threonine 5.4 24 2 U selenocysteine 0.2 1 1 V valine 7.6 34 2 W tryptophan 1.1 5 1 Y tyrosine 1.3 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 selenophosphate synthetase 2 SEPHS1 0.647 selenophosphate synthetase 1 LOC402198 0.276 PREDICTED: similar to hCG1641913 LOC402198 0.276 PREDICTED: similar to hCG1641913 LOC402198 0.273 PREDICTED: similar to hCG1641913 SORCS2 0.014 VPS10 domain receptor protein SORCS 2 FBRS 0.009 fibrosin LOC729660 0.009 PREDICTED: similar to Putative uncharacterized prot... PGR 0.008 progesterone receptor HOXA13 0.008 homeobox A13 LOC731275 0.008 PREDICTED: similar to Putative uncharacterized prot... LOC730036 0.008 PREDICTED: hypothetical protein GSTA5 0.008 glutathione S-transferase alpha 5 RAB11FIP1 0.007 RAB11 family interacting protein 1 isoform 3 RAB11FIP1 0.007 RAB11 family interacting protein 1 isoform 2 COL3A1 0.007 collagen type III alpha 1 preproprotein LOC100293892 0.007 PREDICTED: hypothetical protein GSTA2 0.006 glutathione S-transferase alpha 2 NKX6-1 0.006 NK6 transcription factor related, locus 1 KLHDC7B 0.006 kelch domain containing 7B FAM98C 0.006 hypothetical protein LOC147965 GRIN2D 0.006 N-methyl-D-aspartate receptor subunit 2D precursor ... SCARF1 0.005 scavenger receptor class F, member 1 isoform 1 precu... SCARF1 0.005 scavenger receptor class F, member 1 isoform 5 prec... PGBD4 0.005 piggyBac transposable element derived 4 LOC100287807 0.005 PREDICTED: hypothetical protein LOC100287807 0.005 PREDICTED: hypothetical protein XP_002342309 LOC100293962 0.005 PREDICTED: hypothetical protein LOC100294469 0.005 PREDICTED: hypothetical protein RNF185 0.005 ring finger protein 185 isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
