
| Name: SECISBP2 | Sequence: fasta or formatted (854aa) | NCBI GI: 83779010 | |
|
Description: SECIS binding protein 2
|
Referenced in:
| ||
|
Composition:
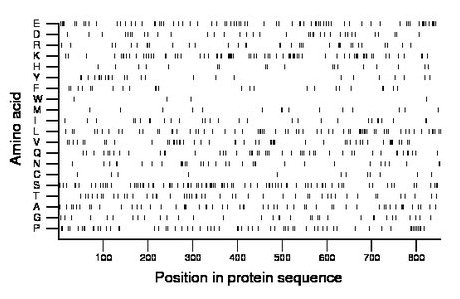
Amino acid Percentage Count Longest homopolymer A alanine 6.7 57 2 C cysteine 1.4 12 1 D aspartate 4.4 38 2 E glutamate 8.7 74 2 F phenylalanine 2.8 24 1 G glycine 4.2 36 2 H histidine 2.2 19 2 I isoleucine 3.7 32 2 K lysine 9.4 80 4 L leucine 7.7 66 2 M methionine 2.0 17 2 N asparagine 4.0 34 2 P proline 7.5 64 2 Q glutamine 5.7 49 2 R arginine 4.3 37 2 S serine 10.2 87 2 T threonine 5.6 48 2 V valine 6.0 51 3 W tryptophan 0.5 4 1 Y tyrosine 2.9 25 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 SECIS binding protein 2 SECISBP2L 0.171 SECIS binding protein 2-like CYLC1 0.014 cylicin, basic protein of sperm head cytoskeleton 1... LOC100131897 0.012 hypothetical protein LOC100131897 NEFH 0.011 neurofilament, heavy polypeptide 200kDa MAP1B 0.010 microtubule-associated protein 1B ASH1L 0.010 absent, small, or homeotic 1-like DEK 0.010 DEK oncogene isoform 2 RIF1 0.010 RAP1 interacting factor 1 EIF5B 0.010 eukaryotic translation initiation factor 5B LOC100133923 0.010 PREDICTED: similar to UPF0634 protein C AP3D1 0.009 adaptor-related protein complex 3, delta 1 subunit ... MUC16 0.009 mucin 16 ZMYND8 0.009 zinc finger, MYND-type containing 8 isoform c ZMYND8 0.009 zinc finger, MYND-type containing 8 isoform a ZMYND8 0.009 zinc finger, MYND-type containing 8 isoform b SHANK2 0.009 SH3 and multiple ankyrin repeat domains 2 isoform 1... DEK 0.008 DEK oncogene isoform 1 FAM133A 0.008 hypothetical protein LOC286499 TCHH 0.008 trichohyalin INCENP 0.008 inner centromere protein antigens 135/155kDa isofor... KIAA1210 0.008 hypothetical protein LOC57481 RB1CC1 0.008 Rb1-inducible coiled coil protein 1 isoform 2 [Homo... RB1CC1 0.008 Rb1-inducible coiled coil protein 1 isoform 1 [Homo... PPP1R12A 0.008 protein phosphatase 1, regulatory (inhibitor) subun... PPP1R12A 0.008 protein phosphatase 1, regulatory (inhibitor) subun... PPP1R12A 0.008 protein phosphatase 1, regulatory (inhibitor) subunit... CIR1 0.007 CBF1 interacting corepressor MYH10 0.007 myosin, heavy polypeptide 10, non-muscle PABPN1 0.007 poly(A) binding protein, nuclear 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
