
| Name: COX19 | Sequence: fasta or formatted (90aa) | NCBI GI: 71979925 | |
|
Description: COX19 cytochrome c oxidase assembly homolog
|
Referenced in:
| ||
|
Composition:
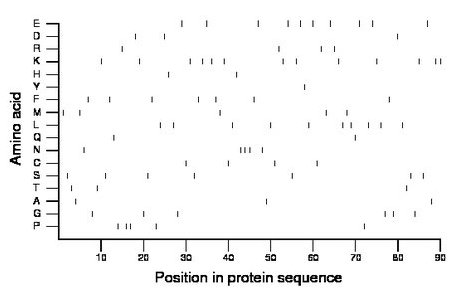
Amino acid Percentage Count Longest homopolymer A alanine 3.3 3 1 C cysteine 4.4 4 1 D aspartate 3.3 3 1 E glutamate 11.1 10 1 F phenylalanine 7.8 7 1 G glycine 6.7 6 1 H histidine 2.2 2 1 I isoleucine 0.0 0 0 K lysine 14.4 13 2 L leucine 11.1 10 1 M methionine 5.6 5 1 N asparagine 5.6 5 3 P proline 5.6 5 2 Q glutamine 2.2 2 1 R arginine 4.4 4 1 S serine 7.8 7 1 T threonine 3.3 3 1 V valine 0.0 0 0 W tryptophan 0.0 0 0 Y tyrosine 1.1 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 COX19 cytochrome c oxidase assembly homolog NDUFS5 0.048 NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa... DEFB119 0.024 defensin, beta 119 isoform a precursor HGS 0.024 hepatocyte growth factor-regulated tyrosine kinase su... HELZ 0.024 helicase with zinc finger domain CHCHD1 0.018 coiled-coil-helix-coiled-coil-helix domain containin... HELQ 0.012 DNA helicase HEL308 ETV6 0.006 ets variant 6 CHCHD3 0.006 coiled-coil-helix-coiled-coil-helix domain containing... RNF157 0.006 ring finger protein 157 LAMA3 0.006 laminin alpha 3 subunit isoform 1 LAMA3 0.006 laminin alpha 3 subunit isoform 4 LAMA3 0.006 laminin alpha 3 subunit isoform 3 LAMA3 0.006 laminin alpha 3 subunit isoform 2 LONRF3 0.006 LON peptidase N-terminal domain and ring finger 3 is... LONRF3 0.006 LON peptidase N-terminal domain and ring finger 3 is... C12orf65 0.006 hypothetical protein LOC91574 C12orf65 0.006 hypothetical protein LOC91574 GLG1 0.006 golgi apparatus protein 1 isoform 3 GLG1 0.006 golgi apparatus protein 1 isoform 2 GLG1 0.006 golgi apparatus protein 1 isoform 1 CHCHD7 0.006 coiled-coil-helix-coiled-coil-helix domain containin... CHCHD7 0.006 coiled-coil-helix-coiled-coil-helix domain containin... CHCHD7 0.006 coiled-coil-helix-coiled-coil-helix domain containin... CYP7B1 0.000 cytochrome P450, family 7, subfamily B, polypeptide 1...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
