
| Name: PRM2 | Sequence: fasta or formatted (102aa) | NCBI GI: 68989267 | |
|
Description: protamine 2
|
Referenced in: Nonhistone Chromosomal Proteins
| ||
|
Composition:
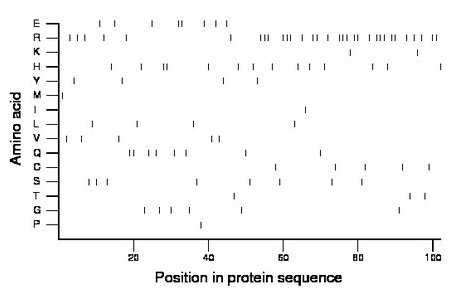
Amino acid Percentage Count Longest homopolymer A alanine 0.0 0 0 C cysteine 4.9 5 1 D aspartate 0.0 0 0 E glutamate 7.8 8 2 F phenylalanine 0.0 0 0 G glycine 5.9 6 1 H histidine 13.7 14 2 I isoleucine 1.0 1 1 K lysine 2.0 2 1 L leucine 3.9 4 1 M methionine 1.0 1 1 N asparagine 0.0 0 0 P proline 1.0 1 1 Q glutamine 7.8 8 2 R arginine 31.4 32 3 S serine 7.8 8 1 T threonine 2.9 3 1 V valine 4.9 5 1 W tryptophan 0.0 0 0 Y tyrosine 3.9 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 protamine 2 PRM1 0.106 protamine 1 RSRC2 0.090 arginine/serine-rich coiled-coil 2 isoform b RSRC2 0.090 arginine/serine-rich coiled-coil 2 isoform a DDX46 0.085 DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 LUC7L2 0.080 LUC7-like 2 C2orf16 0.075 hypothetical protein LOC84226 LUZP4 0.070 leucine zipper protein 4 RBM16 0.065 RNA-binding motif protein 16 SFRS12 0.065 splicing factor, arginine/serine-rich 12 isoform a ... SFRS12 0.065 splicing factor, arginine/serine-rich 12 isoform b [... RBM23 0.055 RNA binding motif protein 23 isoform 1 CROP 0.055 cisplatin resistance-associated overexpressed protei... CROP 0.055 cisplatin resistance-associated overexpressed protei... SNRNP48 0.055 U11/U12 snRNP 48K CDC2L1 0.050 cell division cycle 2-like 1 (PITSLRE proteins) isof... CDC2L1 0.050 cell division cycle 2-like 1 (PITSLRE proteins) isof... CHERP 0.050 calcium homeostasis endoplasmic reticulum protein [... LOC100131202 0.050 PREDICTED: hypothetical protein GPX4 0.050 glutathione peroxidase 4 isoform C precursor LOC100133599 0.050 PREDICTED: hypothetical protein SFRS15 0.045 splicing factor, arginine/serine-rich 15 isoform 3 ... SFRS15 0.045 splicing factor, arginine/serine-rich 15 isoform 2 ... SFRS15 0.045 splicing factor, arginine/serine-rich 15 isoform 1 [... CCNL1 0.045 cyclin L1 CLK1 0.045 CDC-like kinase 1 isoform 2 CLK1 0.045 CDC-like kinase 1 isoform 1 TNIK 0.045 TRAF2 and NCK interacting kinase isoform 8 TNIK 0.045 TRAF2 and NCK interacting kinase isoform 7 TNIK 0.045 TRAF2 and NCK interacting kinase isoform 4Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
