
| Name: AKAP8 | Sequence: fasta or formatted (692aa) | NCBI GI: 5031579 | |
|
Description: A-kinase anchor protein 8
|
Referenced in:
| ||
|
Composition:
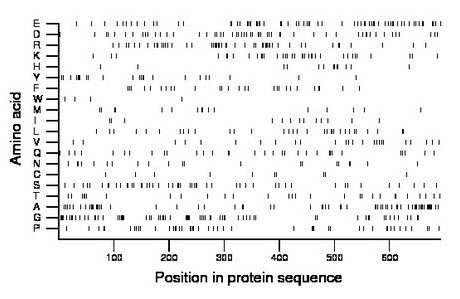
Amino acid Percentage Count Longest homopolymer A alanine 9.5 66 3 C cysteine 1.4 10 1 D aspartate 7.8 54 2 E glutamate 9.7 67 2 F phenylalanine 3.9 27 2 G glycine 10.4 72 4 H histidine 1.9 13 1 I isoleucine 1.9 13 1 K lysine 5.5 38 2 L leucine 4.6 32 2 M methionine 2.5 17 2 N asparagine 3.0 21 2 P proline 6.8 47 3 Q glutamine 4.6 32 1 R arginine 7.5 52 3 S serine 7.2 50 2 T threonine 4.2 29 2 V valine 3.6 25 1 W tryptophan 0.6 4 1 Y tyrosine 3.3 23 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 A-kinase anchor protein 8 AKAP8L 0.196 A kinase (PRKA) anchor protein 8-like ZNF326 0.108 zinc finger protein 326 isoform 1 EIF5B 0.016 eukaryotic translation initiation factor 5B CENPE 0.014 centromere protein E LOC100286959 0.014 PREDICTED: hypothetical protein XP_002343921 BASP1 0.014 brain abundant, membrane attached signal protein 1 [... ZC3H13 0.014 zinc finger CCCH-type containing 13 ERICH1 0.014 glutamate-rich 1 RP1L1 0.013 retinitis pigmentosa 1-like 1 RBM25 0.013 RNA binding motif protein 25 LOC100133599 0.013 PREDICTED: hypothetical protein ZNF131 0.013 zinc finger protein 131 LOC100131202 0.012 PREDICTED: hypothetical protein PRPF38B 0.012 PRP38 pre-mRNA processing factor 38 (yeast) domain ... TCHHL1 0.012 trichohyalin-like 1 CLSPN 0.012 claspin LOC100288837 0.012 PREDICTED: hypothetical protein XP_002343931, parti... SFRS12 0.012 splicing factor, arginine/serine-rich 12 isoform a ... SFRS12 0.012 splicing factor, arginine/serine-rich 12 isoform b [... RSRC2 0.011 arginine/serine-rich coiled-coil 2 isoform a APP 0.011 amyloid beta A4 protein isoform c precursor APP 0.011 amyloid beta A4 protein isoform e precursor MARCKS 0.011 myristoylated alanine-rich protein kinase C substra... FUS 0.011 fusion (involved in t(12;16) in malignant liposarcoma... MPHOSPH10 0.011 M-phase phosphoprotein 10 DIDO1 0.011 death inducer-obliterator 1 isoform c LOC727838 0.010 PREDICTED: similar to cancer/testis antigen family ... SRRT 0.010 arsenate resistance protein 2 isoform e SRRT 0.010 arsenate resistance protein 2 isoform cHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
