
| Name: ERCC4 | Sequence: fasta or formatted (916aa) | NCBI GI: 4885217 | |
|
Description: excision repair cross-complementing rodent repair deficiency, complementation group 4
|
Referenced in: DNases, Recombination, and DNA Repair
| ||
|
Composition:
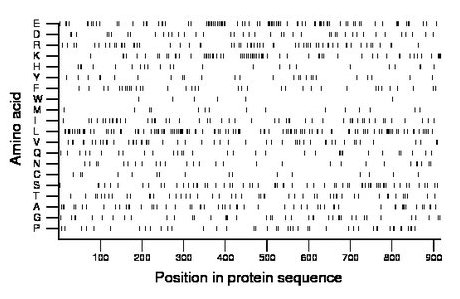
Amino acid Percentage Count Longest homopolymer A alanine 6.4 59 2 C cysteine 1.7 16 1 D aspartate 4.6 42 2 E glutamate 9.9 91 3 F phenylalanine 3.8 35 1 G glycine 4.6 42 2 H histidine 2.5 23 2 I isoleucine 5.6 51 2 K lysine 7.1 65 4 L leucine 12.2 112 2 M methionine 1.7 16 1 N asparagine 3.2 29 2 P proline 4.6 42 1 Q glutamine 3.5 32 2 R arginine 6.8 62 2 S serine 7.1 65 3 T threonine 5.3 49 1 V valine 5.7 52 2 W tryptophan 0.7 6 1 Y tyrosine 2.9 27 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 excision repair cross-complementing rodent repair def... FANCM 0.020 Fanconi anemia, complementation group M LOC100133599 0.009 PREDICTED: hypothetical protein UPF2 0.008 UPF2 regulator of nonsense transcripts homolog [Homo... UPF2 0.008 UPF2 regulator of nonsense transcripts homolog [Homo... TRDN 0.008 triadin ANKRD11 0.008 ankyrin repeat domain 11 CDC42BPA 0.008 CDC42-binding protein kinase alpha isoform B UPF3A 0.007 UPF3 regulator of nonsense transcripts homolog A iso... LOC100133698 0.007 PREDICTED: hypothetical protein LOC100294088 0.007 PREDICTED: hypothetical protein, partial EIF5B 0.007 eukaryotic translation initiation factor 5B MTUS1 0.007 mitochondrial tumor suppressor 1 isoform 1 MTUS1 0.007 mitochondrial tumor suppressor 1 isoform 2 MTUS1 0.007 mitochondrial tumor suppressor 1 isoform 4 LOXHD1 0.007 lipoxygenase homology domains 1 isoform 1 CEP164 0.007 centrosomal protein 164kDa WDR60 0.007 WD repeat domain 60 GOLGA4 0.007 golgi autoantigen, golgin subfamily a, 4 CCDC19 0.007 nasopharyngeal epithelium specific protein 1 LOC283685 0.007 PREDICTED: golgi autoantigen, golgin subfamily a-li... SRRT 0.007 arsenate resistance protein 2 isoform e SRRT 0.007 arsenate resistance protein 2 isoform c CENPE 0.007 centromere protein E BAZ2A 0.007 bromodomain adjacent to zinc finger domain, 2A [Homo... UPF3B 0.006 UPF3 regulator of nonsense transcripts homolog B iso... LOC100133599 0.006 PREDICTED: hypothetical protein BAZ2B 0.006 bromodomain adjacent to zinc finger domain, 2B [Homo... AP3D1 0.006 adaptor-related protein complex 3, delta 1 subunit ... NEFH 0.006 neurofilament, heavy polypeptide 200kDaHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
