
| Name: FANCM | Sequence: fasta or formatted (2048aa) | NCBI GI: 74959747 | |
|
Description: Fanconi anemia, complementation group M
|
Referenced in: DNases, Recombination, and DNA Repair
| ||
|
Composition:
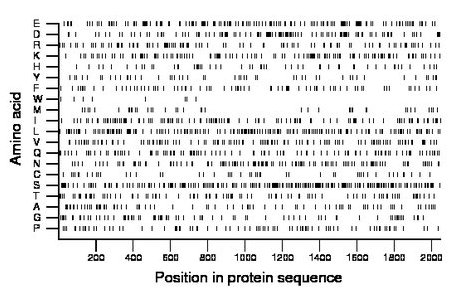
Amino acid Percentage Count Longest homopolymer A alanine 4.1 84 3 C cysteine 2.4 49 1 D aspartate 5.9 120 3 E glutamate 8.2 167 3 F phenylalanine 3.7 76 2 G glycine 4.0 82 2 H histidine 2.6 53 1 I isoleucine 6.0 123 2 K lysine 6.7 138 3 L leucine 8.8 181 3 M methionine 2.1 44 2 N asparagine 6.1 125 2 P proline 4.2 85 3 Q glutamine 5.3 108 2 R arginine 5.2 106 2 S serine 11.2 229 4 T threonine 4.8 98 2 V valine 5.8 118 2 W tryptophan 0.4 9 1 Y tyrosine 2.6 53 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 Fanconi anemia, complementation group M DDX58 0.018 DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide RIG-I [... DHX58 0.012 RNA helicase LGP2 IFIH1 0.011 interferon induced with helicase C domain 1 DICER1 0.010 dicer1 DICER1 0.010 dicer1 ERCC4 0.009 excision repair cross-complementing rodent repair def... DDX27 0.007 DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 DDX21 0.007 DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 EIF4A3 0.006 eukaryotic translation initiation factor 4A, isoform ... DDX50 0.006 nucleolar protein GU2 DDX18 0.006 DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 DDX41 0.005 DEAD-box protein abstrakt CCDC88C 0.005 DVL-binding protein DAPLE LUZP1 0.005 leucine zipper protein 1 LUZP1 0.005 leucine zipper protein 1 IWS1 0.005 IWS1 homolog DDX3X 0.005 DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3 [Homo... DDX3Y 0.005 DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked ... DDX3Y 0.005 DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked [... ASCC3 0.005 activating signal cointegrator 1 complex subunit 3 i... DDX59 0.005 DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 DDX51 0.005 DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 DDX4 0.005 DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 isoform 3 ... DDX4 0.005 DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 isoform 1 [... DDX4 0.005 DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 isoform 2 ... DDX49 0.005 DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 RSF1 0.005 remodeling and spacing factor 1 ASXL3 0.005 additional sex combs like 3 DDX42 0.005 DEAD box polypeptide 42 proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
