
| Name: ANKRD45 | Sequence: fasta or formatted (266aa) | NCBI GI: 38348298 | |
|
Description: ankyrin repeat domain 45
|
Referenced in:
| ||
|
Composition:
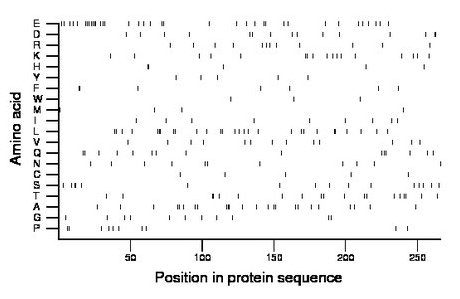
Amino acid Percentage Count Longest homopolymer A alanine 9.0 24 3 C cysteine 1.9 5 1 D aspartate 5.6 15 2 E glutamate 13.9 37 4 F phenylalanine 2.6 7 2 G glycine 4.1 11 1 H histidine 1.9 5 2 I isoleucine 3.4 9 1 K lysine 6.4 17 2 L leucine 10.5 28 3 M methionine 1.5 4 1 N asparagine 4.1 11 1 P proline 3.8 10 2 Q glutamine 5.6 15 2 R arginine 4.5 12 1 S serine 6.0 16 2 T threonine 7.1 19 2 V valine 4.9 13 1 W tryptophan 1.1 3 1 Y tyrosine 1.9 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 ankyrin repeat domain 45 HSPA5 0.069 heat shock 70kDa protein 5 TP53BP2 0.065 tumor protein p53 binding protein, 2 isoform 1 [Hom... TP53BP2 0.065 tumor protein p53 binding protein, 2 isoform 2 ANKRD55 0.063 ankyrin repeat domain 55 isoform 1 PPP1R13B 0.061 apoptosis-stimulating protein of p53, 1 LOC100287718 0.057 hypothetical protein LOC100287718 LOC100293913 0.057 PREDICTED: similar to ankyrin repeat domain 2 [Homo... ANK1 0.057 ankyrin 1 isoform 9 ANK1 0.057 ankyrin 1 isoform 1 ANK1 0.057 ankyrin 1 isoform 3 ANK1 0.057 ankyrin 1 isoform 2 ANK1 0.057 ankyrin 1 isoform 4 ANKRD44 0.053 ankyrin repeat domain 44 ANKS1A 0.051 ankyrin repeat and sterile alpha motif domain conta... ANKDD1A 0.051 ankyrin repeat and death domain containing 1A PSMD10 0.049 proteasome 26S non-ATPase subunit 10 isoform 1 PSMD10 0.049 proteasome 26S non-ATPase subunit 10 isform 2 RIPK4 0.049 ankyrin repeat domain 3 FEM1C 0.049 feminization 1 homolog a NFKBIL2 0.049 NF-kappa-B inhibitor-like protein 2 MYO16 0.047 myosin heavy chain Myr 8 PPP1R12A 0.047 protein phosphatase 1, regulatory (inhibitor) subun... PPP1R12A 0.047 protein phosphatase 1, regulatory (inhibitor) subun... PPP1R12A 0.047 protein phosphatase 1, regulatory (inhibitor) subunit... FEM1B 0.047 fem-1 homolog b ANKRD42 0.047 ankyrin repeat domain 42 PPP1R16B 0.047 protein phosphatase 1 regulatory inhibitor subunit 1... ANK2 0.045 ankyrin 2 isoform 3 ANK2 0.045 ankyrin 2 isoform 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
