
| Name: LOC100293959 | Sequence: fasta or formatted (110aa) | NCBI GI: 239757732 | |
|
Description: PREDICTED: similar to KIAA1654 protein
| Not currently referenced in the text | ||
|
Composition:
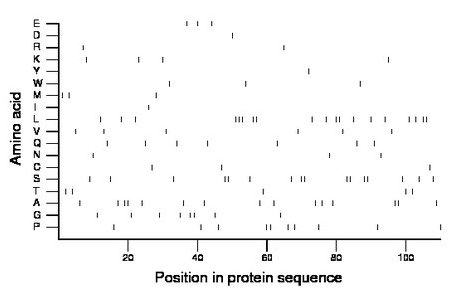
Amino acid Percentage Count Longest homopolymer A alanine 13.6 15 2 C cysteine 2.7 3 1 D aspartate 0.9 1 1 E glutamate 2.7 3 1 F phenylalanine 0.0 0 0 G glycine 7.3 8 2 H histidine 0.0 0 0 I isoleucine 0.9 1 1 K lysine 3.6 4 1 L leucine 17.3 19 3 M methionine 2.7 3 1 N asparagine 2.7 3 1 P proline 9.1 10 2 Q glutamine 6.4 7 1 R arginine 1.8 2 1 S serine 14.5 16 2 T threonine 4.5 5 1 V valine 5.5 6 1 W tryptophan 2.7 3 1 Y tyrosine 0.9 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to KIAA1654 protein CES7 0.041 carboxylesterase 7 isoform 1 CES7 0.041 carboxylesterase 7 isoform 2 GUCY1A2 0.021 guanylate cyclase 1, soluble, alpha 2 CHRNA3 0.021 cholinergic receptor, nicotinic, alpha 3 NOS3 0.015 nitric oxide synthase 3 isoform 4 NOS3 0.015 nitric oxide synthase 3 isoform 3 NOS3 0.015 nitric oxide synthase 3 isoform 2 NOS3 0.015 nitric oxide synthase 3 isoform 1 FAM108A5 0.015 PREDICTED: family with sequence similarity 108, mem... FAM108A5 0.015 PREDICTED: family with sequence similarity 108, mem... FAM108A5 0.015 PREDICTED: hypothetical protein LOC729495 PCK2 0.015 mitochondrial phosphoenolpyruvate carboxykinase 2 is... PCDH20 0.015 protocadherin 20 LOC100293578 0.015 PREDICTED: hypothetical protein LOC100290886 0.015 PREDICTED: hypothetical protein XP_002346949 LOC100286960 0.015 PREDICTED: hypothetical protein XP_002342813 LOC100286960 0.015 PREDICTED: hypothetical protein SRCAP 0.015 Snf2-related CBP activator protein MYO15A 0.010 myosin XV FLJ20021 0.010 PREDICTED: hypothetical protein FLJ20021 0.010 PREDICTED: hypothetical protein FLJ20021 0.010 PREDICTED: hypothetical protein LOC100294236 0.010 PREDICTED: similar to diffuse panbronchiolitis crit... PRAGMIN 0.010 pragmin ADPGK 0.010 ADP-dependent glucokinase MBD6 0.010 methyl-CpG binding domain protein 6 LOC100293844 0.010 PREDICTED: hypothetical protein FAM75A3 0.010 hypothetical protein LOC727830 E2F1 0.005 E2F transcription factor 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
