
| Name: LOC100291805 | Sequence: fasta or formatted (122aa) | NCBI GI: 239757459 | |
|
Description: PREDICTED: similar to Protein FAM27D1
| Not currently referenced in the text | ||
|
Composition:
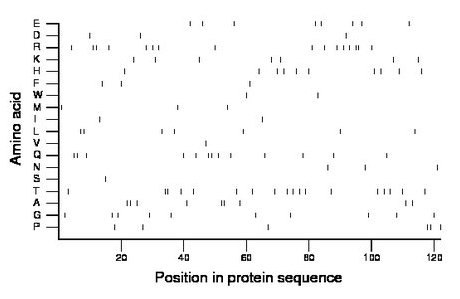
Amino acid Percentage Count Longest homopolymer A alanine 7.4 9 2 C cysteine 0.0 0 0 D aspartate 2.5 3 1 E glutamate 6.6 8 1 F phenylalanine 2.5 3 1 G glycine 8.2 10 1 H histidine 8.2 10 1 I isoleucine 1.6 2 1 K lysine 5.7 7 1 L leucine 5.7 7 2 M methionine 2.5 3 1 N asparagine 2.5 3 1 P proline 4.9 6 2 Q glutamine 10.7 13 2 R arginine 13.1 16 2 S serine 0.8 1 1 T threonine 14.8 18 2 V valine 0.8 1 1 W tryptophan 1.6 2 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to Protein FAM27D1 LOC100289998 1.000 PREDICTED: hypothetical protein XP_002347358 LOC100287921 1.000 PREDICTED: hypothetical protein XP_002344254 LOC100287410 0.786 PREDICTED: hypothetical protein XP_002342959 LOC100287368 0.778 PREDICTED: hypothetical protein XP_002342958 LOC100290475 0.765 PREDICTED: hypothetical protein XP_002347092 LOC100132144 0.748 PREDICTED: similar to Protein FAM27D1 LOC100132859 0.739 PREDICTED: similar to Protein FAM27D1 LOC100293262 0.654 PREDICTED: similar to Protein FAM27D1 LOC100289697 0.654 PREDICTED: hypothetical protein XP_002346920 LOC100287633 0.654 PREDICTED: hypothetical protein XP_002342769 LOC100287633 0.654 PREDICTED: similar to Protein FAM27D1 LOC100130830 0.637 PREDICTED: similar to Protein FAM27E2 LOC100130830 0.637 PREDICTED: similar to Protein FAM27E2 LOC100130830 0.637 PREDICTED: similar to Protein FAM27E2 FAM27E3 0.632 PREDICTED: family with sequence similarity 27, memb... LOC100132144 0.628 PREDICTED: similar to family with sequence similari... LOC100132439 0.620 PREDICTED: similar to Protein FAM27E2 LOC100292121 0.534 PREDICTED: similar to Protein FAM27D1 LOC100289224 0.085 PREDICTED: hypothetical protein XP_002344297 LOC100129488 0.064 PREDICTED: hypothetical protein LOC100129488 0.034 PREDICTED: hypothetical protein LOC100288837 0.034 PREDICTED: hypothetical protein XP_002343931, parti... LOC100286959 0.030 PREDICTED: hypothetical protein XP_002343921 FKBPL 0.030 WAF-1/CIP1 stabilizing protein 39 TOP1 0.026 DNA topoisomerase I CRNN 0.021 cornulin ACIN1 0.021 apoptotic chromatin condensation inducer 1 SFRS18 0.017 splicing factor, arginine/serine-rich 130 SFRS18 0.017 splicing factor, arginine/serine-rich 130Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
