
| Name: LOC100291645 | Sequence: fasta or formatted (345aa) | NCBI GI: 239756555 | |
|
Description: PREDICTED: hypothetical protein XP_002344930
| Not currently referenced in the text | ||
|
Composition:
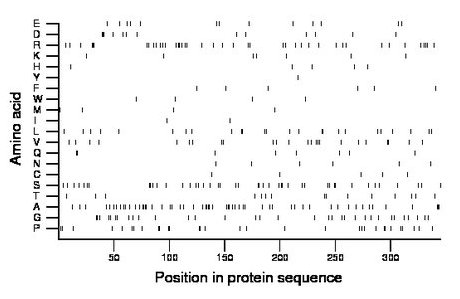
Amino acid Percentage Count Longest homopolymer A alanine 16.5 57 4 C cysteine 1.4 5 1 D aspartate 3.8 13 3 E glutamate 3.8 13 1 F phenylalanine 1.7 6 1 G glycine 9.6 33 2 H histidine 1.4 5 1 I isoleucine 0.6 2 1 K lysine 2.0 7 1 L leucine 7.2 25 2 M methionine 1.2 4 1 N asparagine 1.7 6 1 P proline 9.0 31 2 Q glutamine 2.3 8 2 R arginine 12.2 42 3 S serine 12.8 44 2 T threonine 4.3 15 1 V valine 7.0 24 2 W tryptophan 1.2 4 1 Y tyrosine 0.3 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002344930 LOC100289478 1.000 PREDICTED: hypothetical protein LOC100289478 0.997 PREDICTED: hypothetical protein XP_002343478 SRRM2 0.023 splicing coactivator subunit SRm300 GRIN2D 0.023 N-methyl-D-aspartate receptor subunit 2D precursor ... LOC100292934 0.023 PREDICTED: hypothetical protein GNAS 0.021 GNAS complex locus XLas LOC100133756 0.020 PREDICTED: hypothetical protein, partial ARMCX2 0.020 ALEX2 protein BTBD11 0.020 BTB (POZ) domain containing 11 isoform a ARMCX2 0.020 ALEX2 protein LOC100291912 0.018 PREDICTED: hypothetical protein XP_002344618 LOC100290635 0.018 PREDICTED: hypothetical protein XP_002347290 LOC100287251 0.018 PREDICTED: hypothetical protein XP_002343154 MUC21 0.018 mucin 21 ASPHD1 0.018 aspartate beta-hydroxylase domain containing 1 [Homo... LOC100292082 0.017 PREDICTED: hypothetical protein XP_002345881 FLJ31945 0.017 PREDICTED: hypothetical protein LOC100293254 0.017 PREDICTED: hypothetical protein LOC100289377 0.017 PREDICTED: hypothetical protein LOC100287104 0.017 PREDICTED: hypothetical protein XP_002343630 LOC100289377 0.017 PREDICTED: hypothetical protein XP_002343119 FLJ31945 0.015 PREDICTED: hypothetical protein TMEM88B 0.015 PREDICTED: hypothetical protein LOC643965 FLJ31945 0.015 PREDICTED: hypothetical protein TMEM88B 0.015 PREDICTED: hypothetical protein LOC643965 ANKRD36 0.015 PREDICTED: ankyrin repeat domain 36 MAST2 0.015 microtubule associated serine/threonine kinase 2 [H... TRIO 0.015 triple functional domain (PTPRF interacting) MAP3K12 0.015 mitogen-activated protein kinase kinase kinase 12 [H...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
