
| Name: LOC100291656 | Sequence: fasta or formatted (110aa) | NCBI GI: 239754083 | |
|
Description: PREDICTED: hypothetical protein XP_002345877
| Not currently referenced in the text | ||
|
Composition:
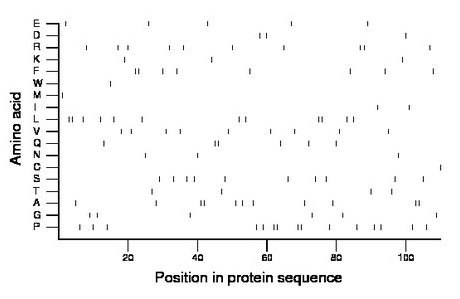
Amino acid Percentage Count Longest homopolymer A alanine 10.0 11 2 C cysteine 0.9 1 1 D aspartate 2.7 3 1 E glutamate 4.5 5 1 F phenylalanine 7.3 8 2 G glycine 5.5 6 1 H histidine 0.0 0 0 I isoleucine 1.8 2 1 K lysine 2.7 3 1 L leucine 10.9 12 2 M methionine 0.9 1 1 N asparagine 2.7 3 1 P proline 13.6 15 2 Q glutamine 5.5 6 2 R arginine 9.1 10 2 S serine 9.1 10 1 T threonine 3.6 4 1 V valine 8.2 9 1 W tryptophan 0.9 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002345877 LOC389328 0.949 PREDICTED: hypothetical protein LOC389328 0.949 PREDICTED: hypothetical protein NUMBL 0.046 numb homolog (Drosophila)-like LOC389151 0.041 hypothetical protein LOC389151 VWF 0.036 von Willebrand factor preproprotein LOC100288993 0.030 PREDICTED: hypothetical protein XP_002342967 ZNF423 0.025 zinc finger protein 423 TANC2 0.025 tetratricopeptide repeat, ankyrin repeat and coiled... BAT2D1 0.020 HBxAg transactivated protein 2 SLC16A11 0.015 solute carrier family 16, member 11 ATP8B2 0.015 ATPase, class I, type 8B, member 2 isoform a PHC1 0.015 polyhomeotic 1-like IQSEC2 0.015 IQ motif and Sec7 domain 2 isoform1 CTPS2 0.015 cytidine triphosphate synthase II CTPS2 0.015 cytidine triphosphate synthase II CTPS2 0.015 cytidine triphosphate synthase II ARHGEF5 0.015 rho guanine nucleotide exchange factor 5 C5orf45 0.015 hypothetical protein LOC51149 isoform 2 C5orf45 0.015 hypothetical protein LOC51149 isoform 1 SALL3 0.010 sal-like 3 GBF1 0.010 golgi-specific brefeldin A resistant guanine nucleoti... SRRM2 0.010 splicing coactivator subunit SRm300 RAVER1 0.010 RAVER1 FOXD1 0.010 forkhead box D1 TTC19 0.010 tetratricopeptide repeat domain 19 FLJ22184 0.010 PREDICTED: hypothetical protein FLJ22184 SGIP1 0.010 SH3-domain GRB2-like (endophilin) interacting prote... HNRNPUL1 0.010 heterogeneous nuclear ribonucleoprotein U-like 1 iso... HNRNPUL1 0.010 heterogeneous nuclear ribonucleoprotein U-like 1 iso...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
