
| Name: LOC100293437 | Sequence: fasta or formatted (203aa) | NCBI GI: 239752933 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
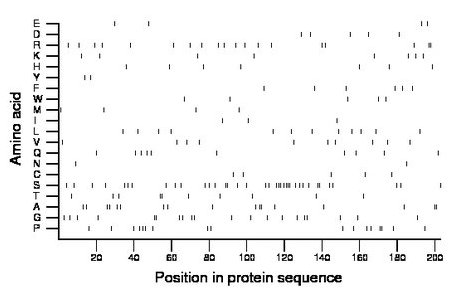
Amino acid Percentage Count Longest homopolymer A alanine 8.4 17 2 C cysteine 2.5 5 1 D aspartate 2.5 5 1 E glutamate 2.0 4 1 F phenylalanine 3.0 6 1 G glycine 9.9 20 2 H histidine 3.4 7 1 I isoleucine 1.5 3 1 K lysine 3.9 8 1 L leucine 5.9 12 1 M methionine 2.0 4 1 N asparagine 1.0 2 1 P proline 8.4 17 2 Q glutamine 4.9 10 1 R arginine 9.4 19 2 S serine 18.2 37 4 T threonine 4.9 10 2 V valine 4.9 10 1 W tryptophan 2.5 5 1 Y tyrosine 1.0 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291237 0.883 PREDICTED: hypothetical protein XP_002347546 LOC100289532 0.883 PREDICTED: hypothetical protein XP_002342183 GREB1 0.033 GREB1 protein isoform a MED1 0.030 mediator complex subunit 1 AKNA 0.030 AT-hook transcription factor SFRS16 0.028 splicing factor, arginine/serine-rich 16 CHD9 0.023 chromodomain helicase DNA binding protein 9 HIC1 0.020 hypermethylated in cancer 1 isoform 2 HIC1 0.020 hypermethylated in cancer 1 isoform 1 MUC6 0.020 mucin 6, gastric SRRM2 0.020 splicing coactivator subunit SRm300 TNRC18 0.020 trinucleotide repeat containing 18 PNN 0.020 pinin, desmosome associated protein SRRM1 0.020 serine/arginine repetitive matrix 1 BAT2 0.018 HLA-B associated transcript-2 LOC100293501 0.018 PREDICTED: hypothetical protein LOC100291178 0.018 PREDICTED: hypothetical protein XP_002347544 LOC100289466 0.018 PREDICTED: hypothetical protein XP_002342181 MACF1 0.018 microfilament and actin filament cross-linker protei... MACF1 0.018 microfilament and actin filament cross-linker protei... CHD7 0.018 chromodomain helicase DNA binding protein 7 MLLT3 0.018 myeloid/lymphoid or mixed-lineage leukemia (trithor... CXorf39 0.018 hypothetical protein LOC139231 MLLT6 0.018 myeloid/lymphoid or mixed-lineage leukemia (trithora... FLJ22184 0.015 PREDICTED: hypothetical protein FLJ22184 LOC100291624 0.015 PREDICTED: hypothetical protein XP_002345876 FLJ22184 0.015 PREDICTED: hypothetical protein FLJ22184 LOC100289712 0.015 PREDICTED: hypothetical protein XP_002346761 LOC100288900 0.015 PREDICTED: hypothetical protein XP_002343772Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
