
| Name: LOC643043 | Sequence: fasta or formatted (61aa) | NCBI GI: 239752299 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {55aa} PREDICTED: hypothetical protein alt prot {55aa} PREDICTED: hypothetical protein | |||
|
Composition:
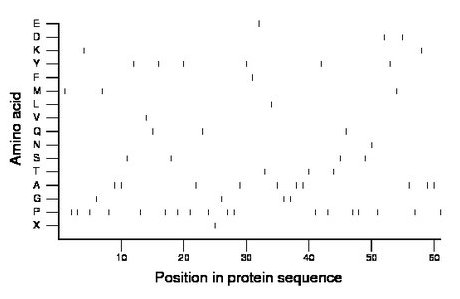
Amino acid Percentage Count Longest homopolymer A alanine 16.4 10 2 C cysteine 0.0 0 0 D aspartate 3.3 2 1 E glutamate 1.6 1 1 F phenylalanine 1.6 1 1 G glycine 6.6 4 2 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 3.3 2 1 L leucine 1.6 1 1 M methionine 4.9 3 1 N asparagine 1.6 1 1 P proline 29.5 18 2 Q glutamine 4.9 3 1 R arginine 0.0 0 0 S serine 6.6 4 1 T threonine 4.9 3 1 V valine 1.6 1 1 W tryptophan 0.0 0 0 X unknown 1.6 1 1 Y tyrosine 9.8 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC643043 0.864 PREDICTED: hypothetical protein LOC643043 0.864 PREDICTED: hypothetical protein SHISA5 0.809 scotin SMARCA2 0.118 SWI/SNF-related matrix-associated actin-dependent re... SMARCA2 0.118 SWI/SNF-related matrix-associated actin-dependent re... HIC1 0.109 hypermethylated in cancer 1 isoform 2 HIC1 0.109 hypermethylated in cancer 1 isoform 1 FUBP1 0.109 far upstream element-binding protein ANXA11 0.100 annexin A11 ANXA11 0.100 annexin A11 ANXA11 0.100 annexin A11 MAPK1IP1L 0.091 MAPK-interacting and spindle-stabilizing protein [Ho... LOC389813 0.091 similar to CG15216-PA SETD1A 0.091 SET domain containing 1A SAMD1 0.091 sterile alpha motif domain containing 1 OGFR 0.082 opioid growth factor receptor SF1 0.082 splicing factor 1 isoform 1 ZFR2 0.082 zinc finger RNA binding protein 2 isoform 1 EIF4G3 0.082 eukaryotic translation initiation factor 4 gamma, 3 ... PRR12 0.082 proline rich 12 CASKIN2 0.082 cask-interacting protein 2 isoform b CASKIN2 0.082 cask-interacting protein 2 isoform a SETD1B 0.073 SET domain containing 1B SFPQ 0.073 splicing factor proline/glutamine rich (polypyrimidin... BCORL1 0.073 BCL6 co-repressor-like 1 EFS 0.073 embryonal Fyn-associated substrate isoform 1 FLJ22184 0.073 PREDICTED: hypothetical protein FLJ22184 CBLL1 0.073 Cas-Br-M (murine) ecotropic retroviral transforming... PRB2 0.073 proline-rich protein BstNI subfamily 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
