
| Name: LOC100289097 | Sequence: fasta or formatted (206aa) | NCBI GI: 239747178 | |
|
Description: PREDICTED: hypothetical protein XP_002343932
| Not currently referenced in the text | ||
|
Composition:
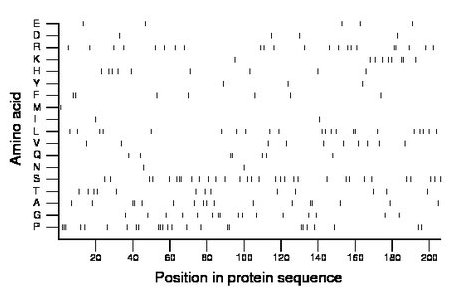
Amino acid Percentage Count Longest homopolymer A alanine 8.3 17 2 C cysteine 0.0 0 0 D aspartate 1.9 4 1 E glutamate 2.4 5 1 F phenylalanine 3.4 7 2 G glycine 7.8 16 2 H histidine 4.4 9 1 I isoleucine 1.0 2 1 K lysine 4.4 9 2 L leucine 10.7 22 2 M methionine 0.5 1 1 N asparagine 1.0 2 1 P proline 12.1 25 3 Q glutamine 3.4 7 2 R arginine 10.7 22 2 S serine 15.5 32 3 T threonine 6.3 13 1 V valine 4.9 10 1 W tryptophan 0.0 0 0 Y tyrosine 1.5 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002343932 CASKIN1 0.039 CASK interacting protein 1 CPSF6 0.039 cleavage and polyadenylation specific factor 6, 68 ... LOC645602 0.036 PREDICTED: hypothetical protein LOC645602 0.036 PREDICTED: hypothetical protein LOC645602 0.036 PREDICTED: hypothetical protein BSN 0.036 bassoon protein PRR12 0.036 proline rich 12 ZFP36 0.034 zinc finger protein 36, C3H type, homolog BRSK1 0.031 BR serine/threonine kinase 1 MAST3 0.031 microtubule associated serine/threonine kinase 3 [H... SHANK1 0.031 SH3 and multiple ankyrin repeat domains 1 MED1 0.031 mediator complex subunit 1 ZNF828 0.028 zinc finger protein 828 MAGEE1 0.028 melanoma antigen family E, 1 AATK 0.026 apoptosis-associated tyrosine kinase HNRNPR 0.026 heterogeneous nuclear ribonucleoprotein R isoform 3... HNRNPR 0.026 heterogeneous nuclear ribonucleoprotein R isoform 1... HNRNPR 0.026 heterogeneous nuclear ribonucleoprotein R isoform 4... HNRNPR 0.026 heterogeneous nuclear ribonucleoprotein R isoform 2 [... DCHS1 0.026 dachsous 1 precursor ZFPM1 0.026 zinc finger protein, multitype 1 HCN2 0.026 hyperpolarization activated cyclic nucleotide-gated... FRG1 0.026 FSHD region gene 1 SRRM2 0.023 splicing coactivator subunit SRm300 RAPH1 0.023 Ras association and pleckstrin homology domains 1 is... ZYX 0.023 zyxin ZYX 0.023 zyxin TACC2 0.023 transforming, acidic coiled-coil containing protein... LOC100133756 0.021 PREDICTED: hypothetical protein, partialHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
