
| Name: LOC100289383 | Sequence: fasta or formatted (149aa) | NCBI GI: 239745658 | |
|
Description: PREDICTED: hypothetical protein XP_002344164
| Not currently referenced in the text | ||
|
Composition:
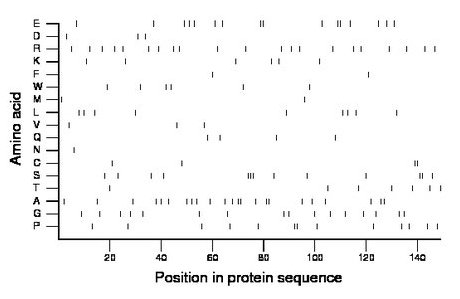
Amino acid Percentage Count Longest homopolymer A alanine 15.4 23 2 C cysteine 2.7 4 2 D aspartate 2.0 3 1 E glutamate 10.7 16 2 F phenylalanine 1.3 2 1 G glycine 10.7 16 1 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 4.0 6 1 L leucine 6.0 9 1 M methionine 1.3 2 1 N asparagine 0.7 1 1 P proline 8.7 13 2 Q glutamine 2.7 4 1 R arginine 14.1 21 1 S serine 8.7 13 3 T threonine 4.7 7 1 V valine 2.0 3 1 W tryptophan 4.0 6 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002344164 LOC100288692 0.962 PREDICTED: hypothetical protein XP_002343084 LOC100294070 0.958 PREDICTED: hypothetical protein LOC100286887 0.955 PREDICTED: hypothetical protein XP_002342631 LOC100288609 0.955 PREDICTED: hypothetical protein XP_002344291 LOC730275 0.566 PREDICTED: hypothetical protein LOC730275 0.566 PREDICTED: hypothetical protein LOC100292758 0.108 PREDICTED: hypothetical protein ZAR1 0.063 zygote arrest 1 CCDC71 0.045 coiled-coil domain containing 71 LOC100129129 0.042 PREDICTED: hypothetical protein PNMAL1 0.035 PNMA-like 1 isoform a ANKRD33B 0.028 PREDICTED: ankyrin repeat domain 33B ANKRD33B 0.028 PREDICTED: ankyrin repeat domain 33B ANKRD33B 0.028 PREDICTED: ankyrin repeat domain 33B C1orf167 0.028 PREDICTED: chromosome 1 open reading frame 167 [Hom... C1orf167 0.028 PREDICTED: chromosome 1 open reading frame 167 [Hom... C1orf167 0.028 PREDICTED: hypothetical protein LOC284498 LOC100287688 0.024 PREDICTED: hypothetical protein XP_002342270 MGC50722 0.024 hypothetical protein LOC399693 LOC100128440 0.024 PREDICTED: hypothetical protein LOC100128440 0.024 PREDICTED: hypothetical protein LOC100128440 0.024 PREDICTED: hypothetical protein SHROOM3 0.024 shroom family member 3 protein PHLDB3 0.024 pleckstrin homology-like domain, family B, member 3... LOC402160 0.024 PREDICTED: similar to hCG18094 LOC651856 0.024 PREDICTED: similar to hCG2023245 KLRG2 0.024 killer cell lectin-like receptor subfamily G, member... FLJ22184 0.021 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.021 PREDICTED: hypothetical protein FLJ22184Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
