
| Name: NUB1 | Sequence: fasta or formatted (601aa) | NCBI GI: 19923477 | |
|
Description: NEDD8 ultimate buster-1
|
Referenced in: Ubiquitin and Related Protein Modifications
| ||
|
Composition:
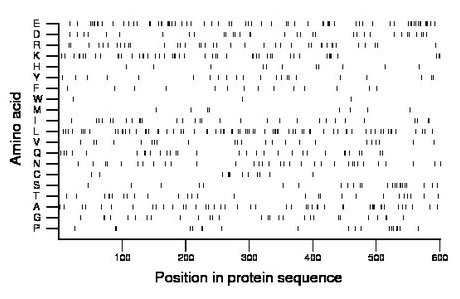
Amino acid Percentage Count Longest homopolymer A alanine 7.7 46 2 C cysteine 2.2 13 2 D aspartate 5.5 33 2 E glutamate 11.1 67 4 F phenylalanine 2.3 14 1 G glycine 4.7 28 2 H histidine 1.8 11 1 I isoleucine 5.2 31 3 K lysine 8.2 49 2 L leucine 12.1 73 2 M methionine 1.5 9 1 N asparagine 5.5 33 1 P proline 3.5 21 2 Q glutamine 5.2 31 2 R arginine 6.7 40 3 S serine 4.2 25 2 T threonine 4.5 27 2 V valine 4.0 24 2 W tryptophan 0.7 4 2 Y tyrosine 3.7 22 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 NEDD8 ultimate buster-1 PPL 0.014 periplakin MYH13 0.012 myosin, heavy polypeptide 13, skeletal muscle [Homo... RAI14 0.012 retinoic acid induced 14 isoform d RAI14 0.012 retinoic acid induced 14 isoform c RAI14 0.012 retinoic acid induced 14 isoform b RAI14 0.012 retinoic acid induced 14 isoform a RAI14 0.012 retinoic acid induced 14 isoform a RAI14 0.012 retinoic acid induced 14 isoform a CCDC150 0.012 coiled-coil domain containing 150 TRIP10 0.011 thyroid hormone receptor interactor 10 GCC2 0.011 GRIP and coiled-coil domain-containing 2 EEA1 0.010 early endosome antigen 1, 162kD ESF1 0.010 ABT1-associated protein CCAR1 0.010 cell-cycle and apoptosis regulatory protein 1 ITSN1 0.010 intersectin 1 isoform ITSN-s ITSN1 0.010 intersectin 1 isoform ITSN-l SCAF1 0.010 SR-related CTD-associated factor 1 C1orf114 0.009 hypothetical protein LOC57821 CCDC68 0.009 coiled-coil domain containing 68 CCDC68 0.009 coiled-coil domain containing 68 PHF14 0.009 PHD finger protein 14 isoform 2 PHF14 0.009 PHD finger protein 14 isoform 1 JAKMIP1 0.008 janus kinase and microtubule interacting protein 1 i... JAKMIP1 0.008 janus kinase and microtubule interacting protein 1 ... LAMA3 0.008 laminin alpha 3 subunit isoform 4 LAMA3 0.008 laminin alpha 3 subunit isoform 3 RB1CC1 0.008 Rb1-inducible coiled coil protein 1 isoform 2 [Homo... RB1CC1 0.008 Rb1-inducible coiled coil protein 1 isoform 1 [Homo... TBC1D17 0.008 TBC1 domain family, member 17Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
