
| Name: LOC100131342 | Sequence: fasta or formatted (197aa) | NCBI GI: 169214652 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {197aa} PREDICTED: hypothetical protein alt mRNA {197aa} PREDICTED: hypothetical protein | |||
|
Composition:
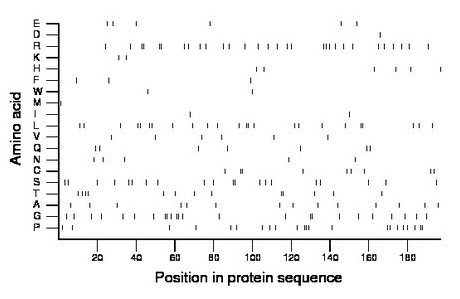
Amino acid Percentage Count Longest homopolymer A alanine 7.1 14 1 C cysteine 4.6 9 2 D aspartate 0.5 1 1 E glutamate 3.0 6 1 F phenylalanine 1.5 3 1 G glycine 12.2 24 2 H histidine 3.0 6 1 I isoleucine 1.0 2 1 K lysine 1.0 2 1 L leucine 12.2 24 2 M methionine 0.5 1 1 N asparagine 2.5 5 1 P proline 11.2 22 3 Q glutamine 3.6 7 1 R arginine 15.2 30 2 S serine 10.2 20 2 T threonine 6.6 13 2 V valine 3.0 6 1 W tryptophan 1.0 2 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100131342 1.000 PREDICTED: hypothetical protein LOC100131342 1.000 PREDICTED: hypothetical protein PHF15 0.034 PHD finger protein 15 LOC100130360 0.031 PREDICTED: hypothetical protein TTBK1 0.028 tau tubulin kinase 1 ZNF444 0.028 zinc finger protein 444 ATP13A2 0.023 ATPase type 13A2 isoform 3 CHD3 0.021 chromodomain helicase DNA binding protein 3 isoform... MAP7D1 0.021 MAP7 domain containing 1 LOC729570 0.021 PREDICTED: hypothetical protein LOC729570 0.021 PREDICTED: hypothetical protein LOC729570 0.021 PREDICTED: hypothetical protein LOC100291750 0.021 PREDICTED: hypothetical protein XP_002345695 ANKRD36 0.021 PREDICTED: ankyrin repeat domain 36 LOC646509 0.021 PREDICTED: hypothetical protein LOC100293536 0.018 PREDICTED: hypothetical protein LOC100291150 0.018 PREDICTED: hypothetical protein XP_002347543 LOC100289439 0.018 PREDICTED: hypothetical protein XP_002342180 SH3RF3 0.018 SH3 domain containing ring finger 3 FOXB1 0.018 forkhead box B1 LOC100291890 0.018 PREDICTED: hypothetical protein XP_002344521 LOC100133756 0.018 PREDICTED: hypothetical protein, partial LOC100289901 0.018 PREDICTED: hypothetical protein XP_002347167 LOC100287920 0.018 PREDICTED: hypothetical protein XP_002344198 LOC100288618 0.018 PREDICTED: hypothetical protein XP_002344285 GPR137 0.018 G protein-coupled receptor 137 SRRM1 0.018 serine/arginine repetitive matrix 1 C18orf56 0.018 hypothetical protein LOC494514 PRICKLE1 0.018 prickle homolog 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
