
| Name: LOC646644 | Sequence: fasta or formatted (129aa) | NCBI GI: 88953955 | |
|
Description: PREDICTED: similar to LLP homolog
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {129aa} PREDICTED: similar to LLP homolog alt mRNA {129aa} PREDICTED: similar to LLP homolog | |||
|
Composition:
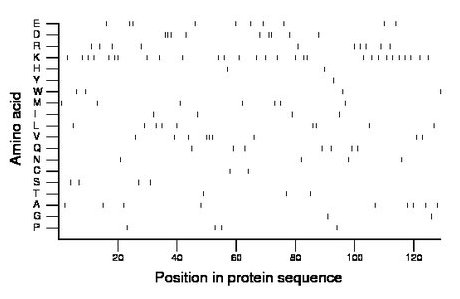
Amino acid Percentage Count Longest homopolymer A alanine 7.0 9 1 C cysteine 1.6 2 1 D aspartate 7.0 9 3 E glutamate 7.8 10 2 F phenylalanine 0.0 0 0 G glycine 1.6 2 1 H histidine 1.6 2 1 I isoleucine 3.1 4 1 K lysine 22.5 29 2 L leucine 7.0 9 2 M methionine 5.4 7 1 N asparagine 3.1 4 1 P proline 3.1 4 1 Q glutamine 5.4 7 1 R arginine 7.8 10 1 S serine 3.1 4 1 T threonine 2.3 3 1 V valine 7.0 9 3 W tryptophan 3.1 4 1 Y tyrosine 0.8 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to LLP homolog LOC646644 1.000 PREDICTED: similar to LLP homolog LOC646644 1.000 PREDICTED: similar to LLP homolog LLPH 0.937 LLP homolog CCDC102B 0.055 coiled-coil domain containing 102B CCDC102B 0.055 coiled-coil domain containing 102B TAF3 0.055 RNA polymerase II transcription factor TAFII140 [Ho... C2orf49 0.046 ashwin C1orf65 0.038 hypothetical protein LOC164127 LRRC45 0.038 leucine rich repeat containing 45 RBBP6 0.038 retinoblastoma-binding protein 6 isoform 2 RBBP6 0.038 retinoblastoma-binding protein 6 isoform 1 LOC100133599 0.034 PREDICTED: hypothetical protein LOC100133599 0.034 PREDICTED: hypothetical protein GOLGA3 0.034 Golgi autoantigen, golgin subfamily a, 3 CCDC121 0.034 coiled-coil domain containing 121 isoform 2 CCDC121 0.034 coiled-coil domain containing 121 isoform 1 CCDC121 0.034 coiled-coil domain containing 121 isoform 3 ANKHD1 0.034 ankyrin repeat and KH domain containing 1 isoform 1 ... ANKHD1-EIF4EBP3 0.034 ANKHD1-EIF4EBP3 protein UTP3 0.034 UTP3, small subunit processome component SMC3 0.029 structural maintenance of chromosomes 3 EIF5B 0.029 eukaryotic translation initiation factor 5B MAN1A2 0.029 mannosidase, alpha, class 1A, member 2 LOC400352 0.029 PREDICTED: similar to Putative golgin subfamily A m... LOC100132816 0.029 PREDICTED: similar to Putative golgin subfamily A m... MYH10 0.029 myosin, heavy polypeptide 10, non-muscle CCDC80 0.029 steroid-sensitive protein 1 CCDC80 0.029 steroid-sensitive protein 1 PPIG 0.029 peptidylprolyl isomerase GHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
