
| Name: EME2 | Sequence: fasta or formatted (444aa) | NCBI GI: 58197552 | |
|
Description: essential meiotic endonuclease 1 homolog 2
|
Referenced in: DNases, Recombination, and DNA Repair
| ||
|
Composition:
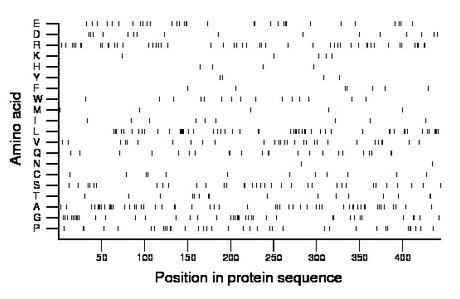
Amino acid Percentage Count Longest homopolymer A alanine 12.2 54 3 C cysteine 2.7 12 1 D aspartate 3.6 16 1 E glutamate 7.2 32 2 F phenylalanine 1.6 7 1 G glycine 9.0 40 3 H histidine 0.9 4 1 I isoleucine 2.0 9 1 K lysine 1.1 5 1 L leucine 11.7 52 5 M methionine 1.1 5 1 N asparagine 0.5 2 1 P proline 8.1 36 2 Q glutamine 5.2 23 2 R arginine 11.0 49 2 S serine 7.7 34 2 T threonine 3.6 16 2 V valine 6.5 29 2 W tryptophan 3.4 15 1 Y tyrosine 0.9 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 essential meiotic endonuclease 1 homolog 2 EME1 0.152 essential meiotic endonuclease 1 homolog 1 MUS81 0.015 MUS81 endonuclease homolog TNRC18 0.014 trinucleotide repeat containing 18 LOC100292157 0.011 PREDICTED: hypothetical protein LOC100290926 0.011 PREDICTED: hypothetical protein XP_002347568 LOC100288995 0.011 PREDICTED: hypothetical protein XP_002343379 LOC100291533 0.011 PREDICTED: hypothetical protein XP_002347080 LOC100288002 0.011 PREDICTED: hypothetical protein XP_002342946 LOC100293375 0.010 PREDICTED: hypothetical protein FBRSL1 0.010 fibrosin-like 1 PTPN21 0.009 protein tyrosine phosphatase, non-receptor type 21 ... LOC100128960 0.008 PREDICTED: similar to paraneoplastic antigen MA2 [H... LOC100128960 0.008 PREDICTED: similar to Putative paraneoplastic antig... SMAD6 0.008 SMAD family member 6 isoform 1 SNAPC3 0.008 small nuclear RNA activating complex, polypeptide 3,... ZNF219 0.008 zinc finger protein 219 ZNF219 0.008 zinc finger protein 219 ZNF219 0.008 zinc finger protein 219 GLRX5 0.007 glutaredoxin 5 BRWD3 0.007 bromodomain and WD repeat domain containing 3 [Homo... LOC100288583 0.007 PREDICTED: hypothetical protein XP_002343504 LOC100129098 0.007 PREDICTED: hypothetical protein MCM3AP 0.007 minichromosome maintenance complex component 3 assoc... C1orf77 0.006 small protein rich in arginine and glycine FLJ31945 0.006 PREDICTED: hypothetical protein LOC100133655 0.006 PREDICTED: hypothetical protein TMPRSS13 0.006 transmembrane protease, serine 13 ZNF579 0.006 zinc finger protein 579 PNMAL2 0.006 PNMA-like 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
