
| Name: GPR143 | Sequence: fasta or formatted (424aa) | NCBI GI: 4557807 | |
|
Description: G protein-coupled receptor 143
|
Referenced in:
| ||
|
Composition:
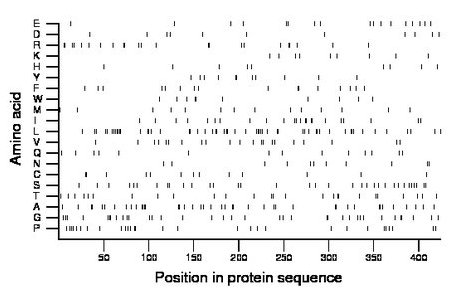
Amino acid Percentage Count Longest homopolymer A alanine 9.9 42 4 C cysteine 3.1 13 2 D aspartate 2.6 11 1 E glutamate 4.0 17 1 F phenylalanine 3.8 16 2 G glycine 8.5 36 2 H histidine 2.1 9 1 I isoleucine 5.4 23 2 K lysine 2.1 9 1 L leucine 13.0 55 3 M methionine 3.3 14 1 N asparagine 2.6 11 1 P proline 6.8 29 2 Q glutamine 3.3 14 1 R arginine 5.4 23 2 S serine 8.0 34 1 T threonine 6.1 26 2 V valine 5.0 21 1 W tryptophan 2.4 10 2 Y tyrosine 2.6 11 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 G protein-coupled receptor 143 VIPR2 0.020 vasoactive intestinal peptide receptor 2 RIMS2 0.017 regulating synaptic membrane exocytosis 2 isoform a... RIMS2 0.017 regulating synaptic membrane exocytosis 2 isoform b... RIMS1 0.011 regulating synaptic membrane exocytosis 1 LOC100132891 0.010 PREDICTED: hypothetical protein LOC100132891 0.010 PREDICTED: hypothetical protein LOC100132891 0.010 PREDICTED: hypothetical protein EMR1 0.010 egf-like module containing, mucin-like, hormone rece... WBP2NL 0.008 WBP2 N-terminal like PHC1 0.008 polyhomeotic 1-like EMR3 0.008 egf-like module-containing mucin-like receptor 3 [H... NFATC4 0.007 nuclear factor of activated T-cells, cytoplasmic, c... NFATC4 0.007 nuclear factor of activated T-cells, cytoplasmic, ca... ATP6V0E2 0.007 ATPase, H+ transporting, V0 subunit isoform 2 [Homo... ATP6V0E2 0.007 ATPase, H+ transporting, V0 subunit isoform 1 [Homo... BDKRB2 0.006 bradykinin receptor B2 ATXN2 0.006 ataxin 2 LOC100292600 0.006 PREDICTED: hypothetical protein LOC100290191 0.006 PREDICTED: hypothetical protein XP_002346744 LOC100289641 0.006 PREDICTED: hypothetical protein XP_002342602 PI4KB 0.006 catalytic phosphatidylinositol 4-kinase beta PTPN23 0.006 protein tyrosine phosphatase, non-receptor type 23 [... GPR157 0.006 G protein-coupled receptor 157 ABCC12 0.006 ATP-binding cassette protein C12 OR2T33 0.006 olfactory receptor, family 2, subfamily T, member 33... DPCR1 0.005 diffuse panbronchiolitis critical region 1 protein ... FAM189B 0.005 hypothetical protein LOC10712 isoform b FAM189B 0.005 hypothetical protein LOC10712 isoform a LOC100129566 0.005 PREDICTED: hypothetical protein LOC100129566Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
