
| Name: HMGA2 | Sequence: fasta or formatted (109aa) | NCBI GI: 4504431 | |
|
Description: high mobility group AT-hook 2 isoform a
|
Referenced in: Adipose Tissue
| ||
Other entries for this name:
alt prot [106aa] high mobility group AT-hook 2 isoform b | |||
|
Composition:
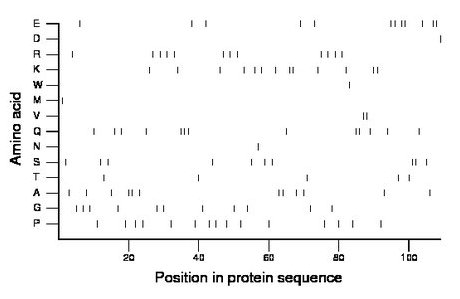
Amino acid Percentage Count Longest homopolymer A alanine 11.0 12 2 C cysteine 0.0 0 0 D aspartate 0.9 1 1 E glutamate 11.0 12 2 F phenylalanine 0.0 0 0 G glycine 10.1 11 1 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 11.9 13 2 L leucine 0.0 0 0 M methionine 0.9 1 1 N asparagine 0.9 1 1 P proline 13.8 15 1 Q glutamine 11.9 13 3 R arginine 11.0 12 1 S serine 9.2 10 2 T threonine 4.6 5 1 V valine 1.8 2 2 W tryptophan 0.9 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 high mobility group AT-hook 2 isoform a HMGA2 0.754 high mobility group AT-hook 2 isoform b HMGA1 0.302 high mobility group AT-hook 1 isoform b HMGA1 0.302 high mobility group AT-hook 1 isoform b HMGA1 0.302 high mobility group AT-hook 1 isoform b HMGA1 0.302 high mobility group AT-hook 1 isoform b HMGA1 0.291 high mobility group AT-hook 1 isoform a HMGA1 0.291 high mobility group AT-hook 1 isoform a LOC100130009 0.256 PREDICTED: hypothetical protein LOC100130009 0.256 PREDICTED: hypothetical protein LOC100130009 0.256 PREDICTED: hypothetical protein C15orf52 0.101 hypothetical protein LOC388115 REXO1 0.085 transcription elongation factor B polypeptide 3 bin... SFRS11 0.085 splicing factor, arginine/serine-rich 11 SYN3 0.085 synapsin III isoform IIIg SYN3 0.085 synapsin III isoform IIIa DOT1L 0.080 DOT1-like, histone H3 methyltransferase PSIP1 0.075 PC4 and SFRS1 interacting protein 1 isoform 2 [Homo... PSIP1 0.075 PC4 and SFRS1 interacting protein 1 isoform 1 [Homo... PSIP1 0.075 PC4 and SFRS1 interacting protein 1 isoform 2 LOC100128074 0.075 PREDICTED: hypothetical protein SYN1 0.075 synapsin I isoform Ia SYN1 0.075 synapsin I isoform Ib HCN2 0.075 hyperpolarization activated cyclic nucleotide-gated... PRB2 0.075 proline-rich protein BstNI subfamily 2 LOC100292370 0.075 PREDICTED: hypothetical protein HS6ST3 0.075 PREDICTED: heparan sulfate 6-O-sulfotransferase 3 [... LOC728650 0.075 PREDICTED: hypothetical protein HS6ST3 0.075 heparan sulfate 6-O-sulfotransferase 3 PRB1 0.075 proline-rich protein BstNI subfamily 1 isoform 1 pre...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
