
| Name: LOC255783 | Sequence: fasta or formatted (142aa) | NCBI GI: 30425446 | |
|
Description: hypothetical protein LOC255783
| Not currently referenced in the text | ||
|
Composition:
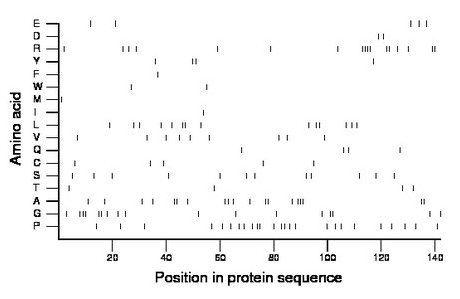
Amino acid Percentage Count Longest homopolymer A alanine 13.4 19 3 C cysteine 3.5 5 1 D aspartate 1.4 2 1 E glutamate 3.5 5 1 F phenylalanine 0.7 1 1 G glycine 12.0 17 3 H histidine 0.0 0 0 I isoleucine 0.7 1 1 K lysine 0.0 0 0 L leucine 9.9 14 2 M methionine 0.7 1 1 N asparagine 0.0 0 0 P proline 17.6 25 2 Q glutamine 2.8 4 1 R arginine 12.0 17 4 S serine 8.5 12 1 T threonine 2.8 4 1 V valine 6.3 9 1 W tryptophan 1.4 2 1 Y tyrosine 2.8 4 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC255783 PRR12 0.062 proline rich 12 TAF4 0.058 TBP-associated factor 4 RLTPR 0.055 RGD motif, leucine rich repeats, tropomodulin domain... RFX1 0.055 regulatory factor X1 GRIN2D 0.055 N-methyl-D-aspartate receptor subunit 2D precursor ... HCN2 0.055 hyperpolarization activated cyclic nucleotide-gated... HOXD11 0.051 homeobox D11 EIF3F 0.051 eukaryotic translation initiation factor 3, subunit 5... WBP11 0.051 WW domain binding protein 11 VASP 0.051 vasodilator-stimulated phosphoprotein UBE2O 0.051 ubiquitin-conjugating enzyme E2O ACR 0.051 acrosin precursor MANEAL 0.051 mannosidase, endo-alpha-like isoform 3 MANEAL 0.051 mannosidase, endo-alpha-like isoform 1 LOC645529 0.051 PREDICTED: similar to Putative acrosin-like proteas... TTLL10 0.051 tubulin tyrosine ligase-like family, member 10 isof... SETD1A 0.047 SET domain containing 1A ATXN2 0.047 ataxin 2 LOC389333 0.047 hypothetical protein LOC389333 MAPK7 0.047 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.047 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.047 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.047 mitogen-activated protein kinase 7 isoform 2 DIAPH2 0.044 diaphanous 2 isoform 12C DIAPH2 0.044 diaphanous 2 isoform 156 NACA 0.044 nascent polypeptide-associated complex alpha subuni... DRD4 0.044 dopamine receptor D4 LOC100132234 0.044 PREDICTED: hypothetical protein LOC100132234 LOC100292723 0.044 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
