
| Name: LOC100292573 | Sequence: fasta or formatted (160aa) | NCBI GI: 239757097 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
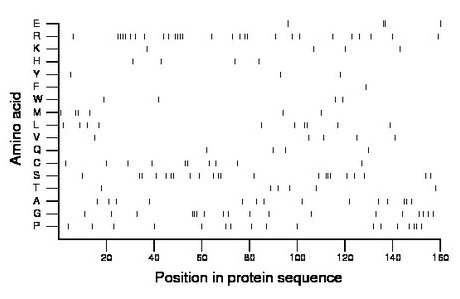
Composition:
Amino acid Percentage Count Longest homopolymer A alanine 8.8 14 2 C cysteine 6.2 10 2 D aspartate 0.0 0 0 E glutamate 2.5 4 2 F phenylalanine 0.6 1 1 G glycine 11.2 18 3 H histidine 2.5 4 1 I isoleucine 0.0 0 0 K lysine 2.5 4 1 L leucine 6.2 10 2 M methionine 3.8 6 2 N asparagine 0.0 0 0 P proline 10.6 17 2 Q glutamine 2.5 4 1 R arginine 17.5 28 4 S serine 13.8 22 3 T threonine 3.8 6 1 V valine 3.1 5 1 W tryptophan 2.5 4 1 Y tyrosine 1.9 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290137 1.000 PREDICTED: hypothetical protein XP_002347927 LOC100287408 1.000 PREDICTED: hypothetical protein XP_002344194 SRRM2 0.037 splicing coactivator subunit SRm300 LOC100293776 0.034 PREDICTED: hypothetical protein LOC100288658 0.034 PREDICTED: hypothetical protein XP_002342750 LOC100288658 0.034 PREDICTED: hypothetical protein LOC100292568 0.031 PREDICTED: hypothetical protein LOC100291146 0.031 PREDICTED: hypothetical protein XP_002346600 LOC100289534 0.031 PREDICTED: hypothetical protein XP_002342442 LOC100293408 0.031 PREDICTED: hypothetical protein LOC100289362 0.031 PREDICTED: hypothetical protein LOC100289362 0.031 PREDICTED: hypothetical protein XP_002343719 LOC100288243 0.028 PREDICTED: hypothetical protein LOC100288243 0.028 PREDICTED: hypothetical protein XP_002342233 CXorf67 0.028 hypothetical protein LOC340602 LOC100292493 0.025 PREDICTED: hypothetical protein LOC100131244 0.025 PREDICTED: similar to kinase D-interacting substrat... LOC100291089 0.025 PREDICTED: hypothetical protein XP_002348116 LOC100131244 0.025 PREDICTED: similar to kinase D-interacting substrat... LOC100287393 0.025 PREDICTED: hypothetical protein XP_002344228 LOC100131244 0.025 PREDICTED: similar to kinase D-interacting substrat... TK2 0.025 thymidine kinase 2, mitochondrial DDN 0.025 dendrin TG 0.022 thyroglobulin LOC729815 0.022 PREDICTED: hypothetical protein GABRA4 0.022 gamma-aminobutyric acid A receptor, alpha 4 precurso... SNRNP27 0.022 small nuclear ribonucleoprotein 27kDa (U4/U6.U5) [Ho... FRAS1 0.022 Fraser syndrome 1 SFRS2B 0.022 splicing factor, arginine/serine-rich 2BHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
