
| Name: LOC100293966 | Sequence: fasta or formatted (307aa) | NCBI GI: 239755208 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
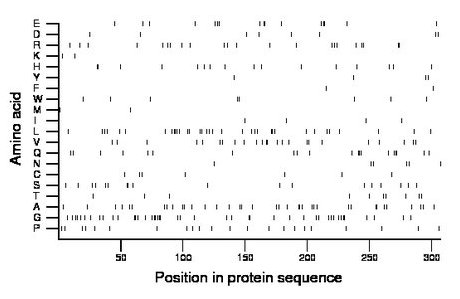
Amino acid Percentage Count Longest homopolymer A alanine 12.1 37 2 C cysteine 2.3 7 1 D aspartate 2.6 8 1 E glutamate 5.2 16 2 F phenylalanine 0.7 2 1 G glycine 14.3 44 3 H histidine 5.2 16 2 I isoleucine 1.3 4 1 K lysine 0.7 2 1 L leucine 11.4 35 3 M methionine 0.7 2 1 N asparagine 2.0 6 1 P proline 7.2 22 1 Q glutamine 6.5 20 3 R arginine 7.2 22 2 S serine 5.9 18 2 T threonine 3.3 10 1 V valine 7.8 24 2 W tryptophan 2.6 8 2 Y tyrosine 1.3 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein CABLES1 0.015 Cdk5 and Abl enzyme substrate 1 isoform 2 LOC284988 0.013 PREDICTED: hypothetical protein COL7A1 0.013 alpha 1 type VII collagen precursor LRRC56 0.011 leucine rich repeat containing 56 LOC100292623 0.011 PREDICTED: hypothetical protein LOC100290254 0.011 PREDICTED: hypothetical protein XP_002347960 COL5A2 0.010 alpha 2 type V collagen preproprotein FUS 0.008 fusion (involved in t(12;16) in malignant liposarcoma... AKR7L 0.008 aflatoxin B1 aldehyde reductase 3 isoform 2 COL6A6 0.008 collagen type VI alpha 6 MMP21 0.008 matrix metalloproteinase 21 preproprotein FAM43B 0.008 family with sequence similarity 43, member B LOC100292572 0.008 PREDICTED: hypothetical protein CHST2 0.008 carbohydrate (N-acetylglucosamine-6-O) sulfotransfer... RASIP1 0.008 Ras-interacting protein 1 C20orf141 0.008 hypothetical protein LOC128653 LOC100129781 0.006 PREDICTED: hypothetical protein ANKS1A 0.006 ankyrin repeat and sterile alpha motif domain conta... LOC100291995 0.006 PREDICTED: hypothetical protein XP_002345272 LOC100293993 0.006 PREDICTED: hypothetical protein LOC100289978 0.006 PREDICTED: hypothetical protein XP_002348035 LOC100289824 0.006 PREDICTED: hypothetical protein XP_002348006 LOC100288302 0.006 PREDICTED: hypothetical protein XP_002343738 LOC100287043 0.006 PREDICTED: hypothetical protein XP_002343726 LOC644246 0.006 PREDICTED: hypothetical protein LOC644246 BPTF 0.006 bromodomain PHD finger transcription factor isoform ... BPTF 0.006 bromodomain PHD finger transcription factor isoform ... COIL 0.006 coilin ARHGAP4 0.006 Rho GTPase activating protein 4Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
