
| Name: LOC100288226 | Sequence: fasta or formatted (154aa) | NCBI GI: 239745424 | |
|
Description: PREDICTED: hypothetical protein XP_002343463
| Not currently referenced in the text | ||
|
Composition:
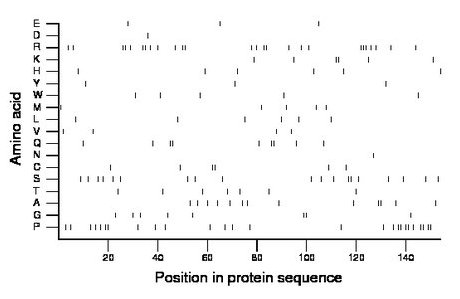
Amino acid Percentage Count Longest homopolymer A alanine 8.4 13 2 C cysteine 3.9 6 2 D aspartate 0.6 1 1 E glutamate 1.9 3 1 F phenylalanine 0.0 0 0 G glycine 5.2 8 2 H histidine 3.9 6 1 I isoleucine 0.0 0 0 K lysine 3.9 6 2 L leucine 3.9 6 1 M methionine 3.2 5 1 N asparagine 0.6 1 1 P proline 16.9 26 2 Q glutamine 5.8 9 2 R arginine 16.9 26 3 S serine 12.3 19 2 T threonine 4.5 7 1 V valine 2.6 4 1 W tryptophan 3.2 5 1 Y tyrosine 1.9 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002343463 BAT2 0.045 HLA-B associated transcript-2 LOC643355 0.042 PREDICTED: hypothetical protein LOC643355 0.042 PREDICTED: hypothetical protein CPEB3 0.038 cytoplasmic polyadenylation element binding protein ... LOC100293644 0.038 PREDICTED: hypothetical protein LOC100287563 0.038 PREDICTED: hypothetical protein LOC100287563 0.038 PREDICTED: hypothetical protein XP_002343733 LOC100132891 0.035 PREDICTED: hypothetical protein LOC100132891 0.035 PREDICTED: hypothetical protein LOC100132891 0.035 PREDICTED: hypothetical protein EMID2 0.035 EMI domain containing 2 LOC729580 0.032 PREDICTED: hypothetical protein LOC729580 0.032 PREDICTED: hypothetical protein LOC729580 0.032 PREDICTED: hypothetical protein LOC100289368 0.032 PREDICTED: hypothetical protein XP_002343360 SRRM2 0.032 splicing coactivator subunit SRm300 KDM6B 0.029 lysine (K)-specific demethylase 6B RAPH1 0.029 Ras association and pleckstrin homology domains 1 is... KCNK4 0.029 TRAAK LOC100291071 0.029 PREDICTED: hypothetical protein XP_002346951 LOC100287198 0.029 PREDICTED: hypothetical protein XP_002342815 LOC100287198 0.029 PREDICTED: hypothetical protein VASP 0.026 vasodilator-stimulated phosphoprotein LOC388906 0.026 PREDICTED: hypothetical protein LOC388906 0.026 PREDICTED: hypothetical protein LOC100290812 0.026 PREDICTED: hypothetical protein XP_002347678 FAM132B 0.026 PREDICTED: family with sequence similarity 132, mem... LOC388906 0.026 PREDICTED: hypothetical protein LOC100288205 0.026 PREDICTED: hypothetical protein XP_002343496Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
