
| Name: LOC100288198 | Sequence: fasta or formatted (105aa) | NCBI GI: 239742921 | |
|
Description: PREDICTED: hypothetical protein XP_002342685
| Not currently referenced in the text | ||
|
Composition:
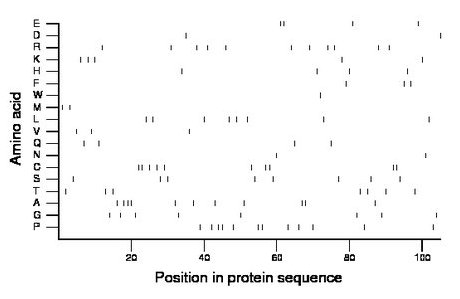
Amino acid Percentage Count Longest homopolymer A alanine 10.5 11 3 C cysteine 9.5 10 2 D aspartate 1.9 2 1 E glutamate 3.8 4 2 F phenylalanine 2.9 3 1 G glycine 7.6 8 1 H histidine 3.8 4 1 I isoleucine 0.0 0 0 K lysine 4.8 5 1 L leucine 7.6 8 1 M methionine 1.9 2 1 N asparagine 1.9 2 1 P proline 11.4 12 2 Q glutamine 3.8 4 1 R arginine 10.5 11 1 S serine 7.6 8 1 T threonine 6.7 7 1 V valine 2.9 3 1 W tryptophan 1.0 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342685 LOC100294043 0.990 PREDICTED: hypothetical protein LOC100290645 0.990 PREDICTED: hypothetical protein XP_002346887 HS3ST3B1 0.039 heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3... ULK1 0.034 Unc-51-like kinase 1 LCP2 0.034 lymphocyte cytosolic protein 2 ALB 0.034 albumin preproprotein BAHCC1 0.024 BAH domain and coiled-coil containing 1 ADAM23 0.024 ADAM metallopeptidase domain 23 preproprotein LOC730130 0.024 hypothetical protein LOC730130 OCEL1 0.024 occludin/ELL domain containing 1 KLF4 0.024 Kruppel-like factor 4 (gut) LOC100291925 0.019 PREDICTED: hypothetical protein XP_002345110 LOC100291392 0.019 PREDICTED: hypothetical protein XP_002347851 LOC100287042 0.019 PREDICTED: hypothetical protein XP_002343584 PPM1J 0.019 protein phosphatase 1J (PP2C domain containing) [Hom... LOC100292095 0.019 PREDICTED: hypothetical protein XP_002345137 LOC100131830 0.019 PREDICTED: hypothetical protein LOC100291303 0.019 PREDICTED: hypothetical protein XP_002347885 LOC100131830 0.019 PREDICTED: hypothetical protein LOC100286950 0.019 PREDICTED: hypothetical protein XP_002344190 LOC100131830 0.019 PREDICTED: hypothetical protein SPINT1 0.019 serine peptidase inhibitor, Kunitz type 1 isoform 2 ... SPINT1 0.019 serine peptidase inhibitor, Kunitz type 1 isoform 2 p... SPINT1 0.019 serine peptidase inhibitor, Kunitz type 1 isoform 1 ... CABLES1 0.019 Cdk5 and Abl enzyme substrate 1 isoform 2 ISL1 0.019 islet-1 GTSE1 0.014 G-2 and S-phase expressed 1 LOC100291743 0.014 PREDICTED: hypothetical protein XP_002344931 LOC100289969 0.014 PREDICTED: hypothetical protein XP_002347692Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
