
| Name: LOC100289470 | Sequence: fasta or formatted (277aa) | NCBI GI: 239742749 | |
|
Description: PREDICTED: hypothetical protein XP_002342626
| Not currently referenced in the text | ||
|
Composition:
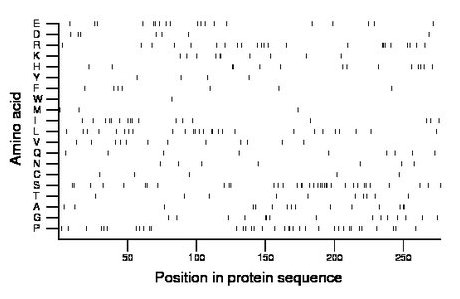
Amino acid Percentage Count Longest homopolymer A alanine 5.4 15 2 C cysteine 1.8 5 1 D aspartate 2.5 7 1 E glutamate 6.9 19 2 F phenylalanine 2.5 7 1 G glycine 5.4 15 2 H histidine 5.1 14 2 I isoleucine 6.1 17 2 K lysine 3.6 10 2 L leucine 9.7 27 2 M methionine 1.1 3 1 N asparagine 2.5 7 1 P proline 13.0 36 2 Q glutamine 3.2 9 1 R arginine 8.3 23 3 S serine 13.0 36 3 T threonine 3.6 10 1 V valine 4.3 12 1 W tryptophan 0.4 1 1 Y tyrosine 1.4 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342626 LOC285679 0.921 hypothetical protein LOC285679 LOC729569 0.801 PREDICTED: hypothetical protein C9orf79 0.208 chromosome 9 open reading frame 79 FAM75A4 0.089 PREDICTED: family with sequence similarity 75, memb... FAM75A6 0.068 hypothetical protein LOC389730 FAM75A2 0.068 hypothetical protein LOC642265 FAM75A3 0.066 hypothetical protein LOC727830 FAM75A1 0.066 hypothetical protein LOC647060 FAM75A5 0.066 hypothetical protein LOC727905 FAM75A7 0.065 hypothetical protein LOC26165 FAM75C1 0.055 family with sequence similarity 75, member C1 [Homo... FAM75A4 0.042 PREDICTED: family with sequence similarity 75, memb... FLJ37078 0.037 hypothetical protein LOC222183 APC2 0.035 adenomatosis polyposis coli 2 LOC100127891 0.033 PREDICTED: similar to hCG2042508 LOC100127891 0.033 PREDICTED: similar to hCG2042508 LOC100127891 0.033 PREDICTED: similar to hCG2042508 LOC100132234 0.031 PREDICTED: hypothetical protein LOC100132234 LOC100132234 0.031 PREDICTED: hypothetical protein LOC100132234 LOC100132234 0.031 PREDICTED: hypothetical protein LOC100132234 SCAF1 0.031 SR-related CTD-associated factor 1 GLTSCR1 0.031 glioma tumor suppressor candidate region gene 1 [Ho... PPP1R13L 0.030 protein phosphatase 1, regulatory subunit 13 like [... PPP1R13L 0.030 protein phosphatase 1, regulatory subunit 13 like [H... MAGEC1 0.030 melanoma antigen family C, 1 WIPF3 0.030 WAS/WASL interacting protein family, member 3 [Homo... MICALL1 0.030 molecule interacting with Rab13 COL27A1 0.028 collagen, type XXVII, alpha 1 PER1 0.028 period 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
