
| Name: MBD3L1 | Sequence: fasta or formatted (194aa) | NCBI GI: 226502212 | |
|
Description: methyl-CpG binding domain protein 3-like
|
Referenced in: DNA Methylation
| ||
|
Composition:
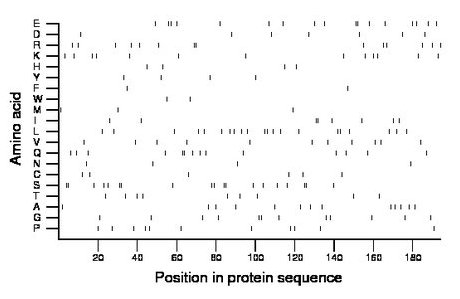
Amino acid Percentage Count Longest homopolymer A alanine 6.7 13 1 C cysteine 3.1 6 1 D aspartate 3.6 7 1 E glutamate 8.8 17 2 F phenylalanine 1.0 2 1 G glycine 6.2 12 2 H histidine 2.1 4 1 I isoleucine 4.1 8 2 K lysine 6.7 13 1 L leucine 10.8 21 2 M methionine 1.5 3 1 N asparagine 2.1 4 1 P proline 5.7 11 1 Q glutamine 7.2 14 2 R arginine 7.2 14 2 S serine 9.8 19 2 T threonine 4.6 9 1 V valine 5.7 11 1 W tryptophan 1.0 2 1 Y tyrosine 2.1 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 methyl-CpG binding domain protein 3-like MBD3 0.337 methyl-CpG binding domain protein 3 MBD2 0.299 methyl-CpG binding domain protein 2 isoform 1 MBD3L3 0.238 PREDICTED: methyl-CpG-binding domain protein 3-like ... MBD3L4 0.238 PREDICTED: methyl-CpG-binding domain protein 3-like ... MBD3L5 0.238 hypothetical protein LOC284428 LOC729458 0.236 PREDICTED: similar to methyl-CpG binding domain pro... MBD3L2 0.236 methyl-CpG binding domain protein 3-like 2 LOC100134199 0.225 PREDICTED: similar to methyl-CpG binding domain pro... HEATR7B2 0.014 HEAT repeat family member 7B2 GRN 0.014 granulin precursor UNKL 0.011 unkempt homolog (Drosophila)-like isoform 1 GTF2IRD1 0.011 GTF2I repeat domain containing 1 isoform 1 GTF2IRD1 0.011 GTF2I repeat domain containing 1 isoform 2 RDH5 0.011 retinol dehydrogenase 5 (11-cis and 9-cis) KIAA0240 0.011 hypothetical protein LOC23506 PODXL2 0.008 podocalyxin-like 2 CCDC52 0.008 coiled-coil domain containing 52 KIAA1632 0.008 hypothetical protein LOC57724 LRRC40 0.008 leucine rich repeat containing 40 UBN1 0.005 ubinuclein 1 UBN1 0.005 ubinuclein 1 ADAM30 0.005 ADAM metallopeptidase domain 30 preproproteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
