
| Name: PHRF1 | Sequence: fasta or formatted (1648aa) | NCBI GI: 221139764 | |
|
Description: PHD and ring finger domains 1
| Not currently referenced in the text | ||
|
Composition:
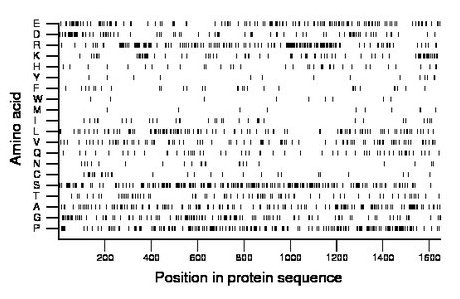
Amino acid Percentage Count Longest homopolymer A alanine 9.1 150 2 C cysteine 1.8 30 1 D aspartate 5.4 89 3 E glutamate 8.3 136 4 F phenylalanine 1.9 31 2 G glycine 6.8 112 3 H histidine 2.2 36 1 I isoleucine 2.2 37 2 K lysine 5.0 83 4 L leucine 6.7 111 3 M methionine 0.7 11 1 N asparagine 1.5 24 1 P proline 10.6 174 3 Q glutamine 3.2 53 1 R arginine 10.5 173 3 S serine 12.6 208 3 T threonine 4.4 73 2 V valine 5.6 93 2 W tryptophan 0.5 9 1 Y tyrosine 0.9 15 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PHD and ring finger domains 1 SCAF1 0.046 SR-related CTD-associated factor 1 SFRS2IP 0.031 splicing factor, arginine/serine-rich 2, interactin... SRRM2 0.024 splicing coactivator subunit SRm300 LOC100170229 0.024 hypothetical protein LOC100170229 SON 0.022 SON DNA-binding protein isoform F SON 0.022 SON DNA-binding protein isoform B SFRS4 0.020 splicing factor, arginine/serine-rich 4 RSRC2 0.020 arginine/serine-rich coiled-coil 2 isoform a SFRS12 0.018 splicing factor, arginine/serine-rich 12 isoform a ... SFRS12 0.018 splicing factor, arginine/serine-rich 12 isoform b [... PPIG 0.018 peptidylprolyl isomerase G PRPF38B 0.017 PRP38 pre-mRNA processing factor 38 (yeast) domain ... LOC100133599 0.016 PREDICTED: hypothetical protein SRRM1 0.016 serine/arginine repetitive matrix 1 GPATCH8 0.016 G patch domain containing 8 CROP 0.016 cisplatin resistance-associated overexpressed protei... CROP 0.016 cisplatin resistance-associated overexpressed protei... LOC100286959 0.015 PREDICTED: hypothetical protein XP_002343921 RSRC1 0.015 arginine/serine-rich coiled-coil 1 SFRS11 0.015 splicing factor, arginine/serine-rich 11 DDX46 0.015 DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 KDM5B 0.015 jumonji, AT rich interactive domain 1B LOC100133760 0.015 PREDICTED: similar to Jumonji, AT rich interactive ... SFRS8 0.015 splicing factor, arginine/serine-rich 8 CDC2L5 0.015 cell division cycle 2-like 5 isoform 1 CDC2L5 0.015 cell division cycle 2-like 5 isoform 2 ACIN1 0.014 apoptotic chromatin condensation inducer 1 KDM5A 0.014 retinoblastoma binding protein 2 isoform 1 KDM5A 0.014 retinoblastoma binding protein 2 isoform 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
