
| Name: FAM134A | Sequence: fasta or formatted (543aa) | NCBI GI: 187607445 | |
|
Description: hypothetical protein LOC79137
| Not currently referenced in the text | ||
|
Composition:
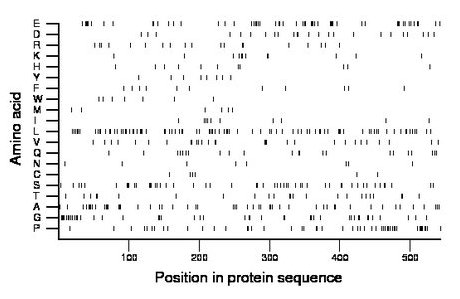
Amino acid Percentage Count Longest homopolymer A alanine 9.4 51 3 C cysteine 1.1 6 1 D aspartate 4.4 24 1 E glutamate 10.5 57 5 F phenylalanine 2.2 12 2 G glycine 8.5 46 5 H histidine 2.2 12 2 I isoleucine 1.8 10 1 K lysine 2.2 12 2 L leucine 15.8 86 2 M methionine 1.3 7 1 N asparagine 1.5 8 1 P proline 9.4 51 2 Q glutamine 4.1 22 2 R arginine 3.5 19 1 S serine 9.6 52 4 T threonine 4.4 24 2 V valine 5.3 29 2 W tryptophan 1.3 7 1 Y tyrosine 1.5 8 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC79137 FAM134C 0.196 hypothetical protein LOC162427 FAM134B 0.140 hypothetical protein LOC54463 isoform 1 FAM134B 0.114 hypothetical protein LOC54463 isoform 2 MLL2 0.027 myeloid/lymphoid or mixed-lineage leukemia 2 RERE 0.023 atrophin-1 like protein isoform b RERE 0.023 atrophin-1 like protein isoform a RERE 0.023 atrophin-1 like protein isoform a FBLIM1 0.022 filamin-binding LIM protein-1 isoform b FBLIM1 0.022 filamin-binding LIM protein-1 isoform a CDGAP 0.021 Cdc42 GTPase-activating protein SETD1A 0.020 SET domain containing 1A NEFM 0.020 neurofilament, medium polypeptide 150kDa isoform 1 ... NEFM 0.020 neurofilament, medium polypeptide 150kDa isoform 2 ... COL15A1 0.019 alpha 1 type XV collagen precursor FLJ22184 0.018 PREDICTED: hypothetical protein FLJ22184 ARAP1 0.018 ArfGAP with RhoGAP domain, ankyrin repeat and PH dom... PRR12 0.018 proline rich 12 MBD6 0.017 methyl-CpG binding domain protein 6 FAM75A6 0.016 hypothetical protein LOC389730 FAM75A2 0.016 hypothetical protein LOC642265 FAM75A1 0.016 hypothetical protein LOC647060 ARL6IP1 0.016 ADP-ribosylation factor-like 6 interacting protein [... FAM75A4 0.016 PREDICTED: family with sequence similarity 75, memb... FAM75A4 0.016 PREDICTED: family with sequence similarity 75, memb... FAM75A7 0.016 hypothetical protein LOC26165 FAM75A3 0.016 hypothetical protein LOC727830 FAM75A5 0.016 hypothetical protein LOC727905 FMNL1 0.015 formin-like 1 MUC16 0.015 mucin 16Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
