
| Name: NOL3 | Sequence: fasta or formatted (208aa) | NCBI GI: 4505419 | |
|
Description: nucleolar protein 3
|
Referenced in:
| ||
|
Composition:
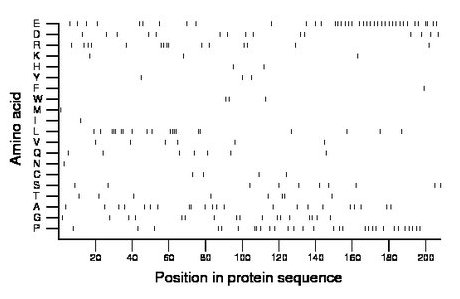
Amino acid Percentage Count Longest homopolymer A alanine 10.6 22 2 C cysteine 1.9 4 1 D aspartate 7.2 15 1 E glutamate 18.8 39 2 F phenylalanine 0.5 1 1 G glycine 8.2 17 2 H histidine 1.0 2 1 I isoleucine 0.5 1 1 K lysine 1.4 3 1 L leucine 9.6 20 4 M methionine 0.5 1 1 N asparagine 0.5 1 1 P proline 14.4 30 2 Q glutamine 3.4 7 1 R arginine 7.2 15 2 S serine 4.8 10 1 T threonine 3.8 8 2 V valine 2.9 6 1 W tryptophan 1.4 3 1 Y tyrosine 1.4 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 nucleolar protein 3 CARD6 0.127 caspase recruitment domain family, member 6 SLC16A2 0.090 solute carrier family 16, member 2 DGKK 0.080 diacylglycerol kinase kappa TRIM44 0.078 DIPB protein RYR1 0.078 skeletal muscle ryanodine receptor isoform 2 RYR1 0.078 skeletal muscle ryanodine receptor isoform 1 RP1L1 0.073 retinitis pigmentosa 1-like 1 SLC26A8 0.063 solute carrier family 26, member 8 isoform a SLC26A8 0.063 solute carrier family 26, member 8 isoform b PDGFRB 0.061 platelet-derived growth factor receptor beta precurso... PRRT2 0.061 proline-rich transmembrane protein 2 PPM1E 0.058 protein phosphatase 1E VSIG1 0.058 V-set and immunoglobulin domain containing 1 BASP1 0.056 brain abundant, membrane attached signal protein 1 [... OGFR 0.056 opioid growth factor receptor HCLS1 0.056 hematopoietic cell-specific Lyn substrate 1 ZNF219 0.051 zinc finger protein 219 ZNF219 0.051 zinc finger protein 219 ZNF219 0.051 zinc finger protein 219 ACAN 0.051 aggrecan isoform 2 precursor ACAN 0.051 aggrecan isoform 1 precursor CCNB1 0.051 cyclin B1 PHLDA1 0.051 pleckstrin homology-like domain, family A, member 1 ... SULT2B1 0.049 sulfotransferase family, cytosolic, 2B, member 1 iso... SULT2B1 0.049 sulfotransferase family, cytosolic, 2B, member 1 iso... ZNF358 0.049 zinc finger protein 358 PRDM2 0.049 retinoblastoma protein-binding zinc finger protein i... PRDM2 0.049 retinoblastoma protein-binding zinc finger protein i... CLIC6 0.049 chloride intracellular channel 6Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
