
| Name: HCCA2 | Sequence: fasta or formatted (237aa) | NCBI GI: 34594669 | |
|
Description: HCCA2 protein
|
Referenced in: Spindle, M phase, and Meiosis
| ||
|
Composition:
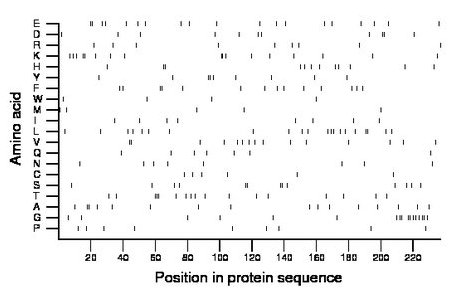
Amino acid Percentage Count Longest homopolymer A alanine 6.8 16 2 C cysteine 2.5 6 1 D aspartate 4.6 11 2 E glutamate 8.0 19 2 F phenylalanine 5.9 14 2 G glycine 7.2 17 2 H histidine 5.1 12 2 I isoleucine 3.4 8 1 K lysine 7.2 17 2 L leucine 8.9 21 2 M methionine 2.1 5 1 N asparagine 3.4 8 1 P proline 3.8 9 1 Q glutamine 3.0 7 1 R arginine 3.8 9 1 S serine 5.5 13 2 T threonine 7.2 17 3 V valine 6.3 15 2 W tryptophan 1.7 4 1 Y tyrosine 3.8 9 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 HCCA2 protein MOBKL1A 0.296 MOB1, Mps One Binder kinase activator-like 1A MOBKL1B 0.292 Mob4B protein MOBKL2C 0.224 MOB1, Mps One Binder kinase activator-like 2C isofor... MOBKL2C 0.224 MOB1, Mps One Binder kinase activator-like 2C isofor... MOBKL2A 0.222 MOB-LAK MOBKL2B 0.214 MOB1, Mps One Binder kinase activator-like 2B MOBKL3 0.023 Mps One Binder kinase activator-like 3 isoform 3 [H... MOBKL3 0.023 Mps One Binder kinase activator-like 3 isoform 2 [Ho... MOBKL3 0.023 Mps One Binder kinase activator-like 3 isoform 1 [Ho... TRIO 0.017 triple functional domain (PTPRF interacting) FZD8 0.017 frizzled 8 C2orf29 0.017 hypothetical protein LOC55571 YEATS2 0.015 YEATS domain containing 2 TRIM67 0.015 tripartite motif-containing 67 KRT9 0.013 keratin 9 SLC4A5 0.013 sodium bicarbonate transporter 4 isoform a SLC4A5 0.013 sodium bicarbonate transporter 4 isoform c ZIC2 0.013 zinc finger protein of the cerebellum 2 ANKS1A 0.013 ankyrin repeat and sterile alpha motif domain conta... ZFHX3 0.013 AT-binding transcription factor 1 GATAD1 0.013 GATA zinc finger domain containing 1 FNDC3B 0.013 fibronectin type III domain containing 3B FNDC3B 0.013 fibronectin type III domain containing 3B CAPNS1 0.013 calpain, small subunit 1 CAPNS1 0.013 calpain, small subunit 1 ARID1B 0.013 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.013 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.013 AT rich interactive domain 1B (SWI1-like) isoform 3 ... LOC339192 0.013 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
