
| Name: LOC100293495 | Sequence: fasta or formatted (192aa) | NCBI GI: 239753821 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
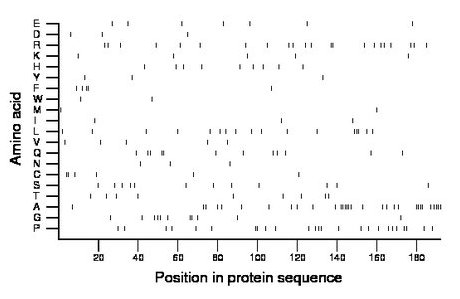
Amino acid Percentage Count Longest homopolymer A alanine 16.1 31 6 C cysteine 3.1 6 2 D aspartate 1.6 3 1 E glutamate 3.6 7 1 F phenylalanine 2.6 5 2 G glycine 5.7 11 2 H histidine 4.7 9 1 I isoleucine 1.6 3 1 K lysine 2.6 5 1 L leucine 8.9 17 3 M methionine 1.0 2 1 N asparagine 1.6 3 1 P proline 13.0 25 2 Q glutamine 6.2 12 2 R arginine 11.5 22 2 S serine 6.2 12 1 T threonine 4.7 9 1 V valine 2.6 5 1 W tryptophan 1.0 2 1 Y tyrosine 1.6 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291075 0.053 PREDICTED: hypothetical protein XP_002346651 LOC100288832 0.053 PREDICTED: hypothetical protein XP_002342488 RTN4 0.042 reticulon 4 isoform B RTN4 0.042 reticulon 4 isoform D RTN4 0.042 reticulon 4 isoform A SAMD1 0.040 sterile alpha motif domain containing 1 FAM48B1 0.037 hypothetical protein LOC100130302 CTNND2 0.034 catenin (cadherin-associated protein), delta 2 (neur... LOC100292048 0.032 PREDICTED: hypothetical protein XP_002344877 LOC100293769 0.032 PREDICTED: hypothetical protein LOC100291339 0.032 PREDICTED: hypothetical protein XP_002347364 LOC100287626 0.032 PREDICTED: hypothetical protein XP_002343250 CDKN1C 0.029 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.029 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.029 cyclin-dependent kinase inhibitor 1C isoform a LOC645317 0.029 PREDICTED: similar to coiled-coil-helix-coiled-coil... LOC645317 0.029 PREDICTED: similar to coiled-coil-helix-coiled-coil... LOC645317 0.029 PREDICTED: similar to coiled-coil-helix-coiled-coil... NLK 0.029 nemo like kinase IER5L 0.029 immediate early response 5-like LOC100291579 0.027 PREDICTED: hypothetical protein XP_002345911 FLJ22184 0.027 PREDICTED: hypothetical protein FLJ22184 GGN 0.027 gametogenetin KLF16 0.027 BTE-binding protein 4 SRCAP 0.027 Snf2-related CBP activator protein LOC100289126 0.027 PREDICTED: hypothetical protein XP_002343381 AMOT 0.027 angiomotin isoform 2 AMOT 0.027 angiomotin isoform 1 SCARF2 0.027 scavenger receptor class F, member 2 isoform 1 [Homo...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
