
| Name: LOC100288980 | Sequence: fasta or formatted (281aa) | NCBI GI: 239746757 | |
|
Description: PREDICTED: hypothetical protein XP_002343835
| Not currently referenced in the text | ||
|
Composition:
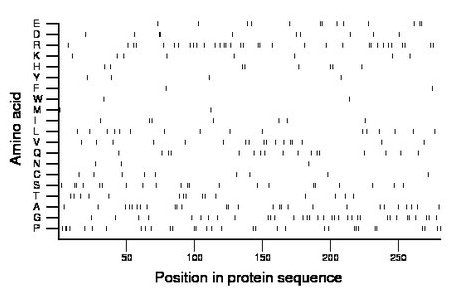
Amino acid Percentage Count Longest homopolymer A alanine 9.6 27 2 C cysteine 3.2 9 1 D aspartate 3.6 10 2 E glutamate 4.6 13 2 F phenylalanine 0.7 2 1 G glycine 12.1 34 2 H histidine 2.8 8 1 I isoleucine 2.5 7 1 K lysine 2.8 8 1 L leucine 5.7 16 1 M methionine 0.7 2 1 N asparagine 1.1 3 1 P proline 11.4 32 2 Q glutamine 6.0 17 1 R arginine 12.1 34 2 S serine 7.5 21 2 T threonine 5.3 15 1 V valine 6.0 17 2 W tryptophan 0.7 2 1 Y tyrosine 1.4 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002343835 LOC100289935 0.692 PREDICTED: hypothetical protein XP_002348168 LOC100292402 0.528 PREDICTED: hypothetical protein ANKRD33B 0.021 PREDICTED: ankyrin repeat domain 33B LOC728767 0.021 PREDICTED: hypothetical protein LOC728767 0.021 PREDICTED: hypothetical protein LOC100289368 0.021 PREDICTED: hypothetical protein XP_002343360 LOC100293975 0.016 PREDICTED: hypothetical protein JPH2 0.016 junctophilin 2 isoform 1 FLJ31945 0.014 PREDICTED: hypothetical protein LOC100290812 0.014 PREDICTED: hypothetical protein XP_002347678 FLJ31945 0.014 PREDICTED: hypothetical protein LOC100288205 0.014 PREDICTED: hypothetical protein XP_002343496 FLJ31945 0.014 PREDICTED: hypothetical protein C6orf132 0.012 PREDICTED: chromosome 6 open reading frame 132 [Hom... LOC100292133 0.012 PREDICTED: hypothetical protein LOC100291976 0.012 PREDICTED: hypothetical protein XP_002345754 C6orf132 0.012 PREDICTED: chromosome 6 open reading frame 132 [Hom... LOC100294354 0.012 PREDICTED: hypothetical protein C6orf132 0.012 PREDICTED: hypothetical protein LOC647024 GRK6 0.012 G protein-coupled receptor kinase 6 isoform B BCL9L 0.012 B-cell CLL/lymphoma 9-like LOC389174 0.012 PREDICTED: hypothetical protein LOC100291718 0.012 PREDICTED: hypothetical protein XP_002345751 LOC100287726 0.012 PREDICTED: hypothetical protein LOC389174 0.012 PREDICTED: hypothetical protein LOC100287726 0.012 PREDICTED: hypothetical protein XP_002342449 LOC389174 0.012 PREDICTED: hypothetical protein LENG1 0.012 leukocyte receptor cluster (LRC) member 1 LOC100129203 0.012 PREDICTED: similar to mCG50504Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
