
| Name: LOC100287204 | Sequence: fasta or formatted (161aa) | NCBI GI: 239746315 | |
|
Description: PREDICTED: hypothetical protein XP_002343693
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {161aa} PREDICTED: hypothetical protein | |||
|
Composition:
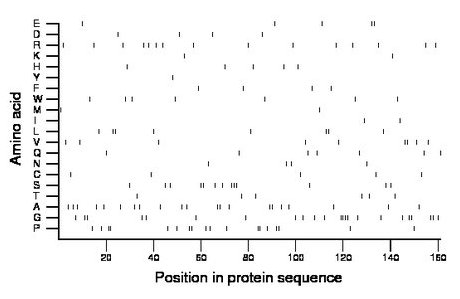
Amino acid Percentage Count Longest homopolymer A alanine 13.0 21 2 C cysteine 3.1 5 1 D aspartate 2.5 4 1 E glutamate 3.1 5 2 F phenylalanine 2.5 4 1 G glycine 14.3 23 4 H histidine 3.1 5 1 I isoleucine 1.2 2 1 K lysine 1.2 2 1 L leucine 5.0 8 2 M methionine 1.2 2 1 N asparagine 2.5 4 1 P proline 11.2 18 2 Q glutamine 4.3 7 1 R arginine 9.3 15 1 S serine 8.1 13 3 T threonine 3.7 6 1 V valine 5.6 9 2 W tryptophan 4.3 7 1 Y tyrosine 0.6 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002343693 LOC100287204 1.000 PREDICTED: hypothetical protein LOC100289763 0.049 PREDICTED: hypothetical protein XP_002347867 LOC100289681 0.049 PREDICTED: hypothetical protein XP_002343623 LOC100292172 0.046 PREDICTED: hypothetical protein LOC100292217 0.034 PREDICTED: hypothetical protein LOC100288033 0.034 PREDICTED: hypothetical protein LOC100288033 0.034 PREDICTED: hypothetical protein XP_002343859 LOC644794 0.024 PREDICTED: hypothetical protein LOC644794 0.024 PREDICTED: hypothetical protein LOC644794 0.024 PREDICTED: hypothetical protein LOC644794 0.024 PREDICTED: hypothetical protein TBKBP1 0.024 TBK1 binding protein 1 C6orf136 0.021 hypothetical protein LOC221545 isoform 3 CDC42EP1 0.021 CDC42 effector protein 1 FAM100B 0.021 hypothetical protein LOC283991 KCNH3 0.021 potassium voltage-gated channel, subfamily H (eag-re... RTN4 0.021 reticulon 4 isoform D RTN4 0.021 reticulon 4 isoform A LOC100293578 0.021 PREDICTED: hypothetical protein LOC100290886 0.021 PREDICTED: hypothetical protein XP_002346949 LOC100286960 0.021 PREDICTED: hypothetical protein XP_002342813 LOC100286960 0.021 PREDICTED: hypothetical protein PLEKHA4 0.021 pleckstrin homology domain containing family A memb... PLEKHA4 0.021 pleckstrin homology domain containing family A memb... KIAA0323 0.018 hypothetical protein LOC23351 LMOD2 0.018 leiomodin 2 (cardiac) CXorf49 0.015 hypothetical protein LOC100130361 CXorf49B 0.015 hypothetical protein LOC100132994 RTN4 0.015 reticulon 4 isoform BHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
