
| Name: LOC100287449 | Sequence: fasta or formatted (146aa) | NCBI GI: 239745689 | |
|
Description: PREDICTED: hypothetical protein XP_002343527
| Not currently referenced in the text | ||
|
Composition:
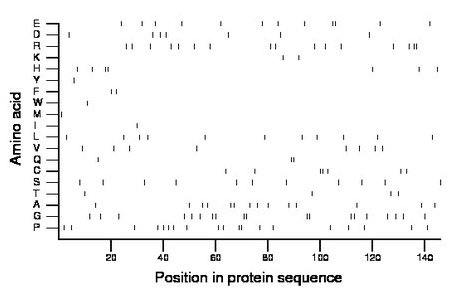
Amino acid Percentage Count Longest homopolymer A alanine 9.6 14 2 C cysteine 4.8 7 2 D aspartate 4.8 7 1 E glutamate 8.2 12 2 F phenylalanine 1.4 2 1 G glycine 13.0 19 2 H histidine 4.8 7 2 I isoleucine 0.7 1 1 K lysine 1.4 2 1 L leucine 7.5 11 1 M methionine 0.7 1 1 N asparagine 0.0 0 0 P proline 13.0 19 2 Q glutamine 2.1 3 2 R arginine 11.0 16 2 S serine 7.5 11 1 T threonine 2.7 4 1 V valine 5.5 8 1 W tryptophan 0.7 1 1 Y tyrosine 0.7 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002343527 LOC100292020 1.000 PREDICTED: similar to NLRP1 protein LOC100291139 1.000 PREDICTED: hypothetical protein XP_002347754 USP51 0.045 ubiquitin specific protease 51 LOC729370 0.038 PREDICTED: hypothetical protein KLHL17 0.038 kelch-like 17 MAGI3 0.034 membrane-associated guanylate kinase-related 3 iso... MAGI3 0.034 membrane-associated guanylate kinase-related 3 iso... HCN2 0.034 hyperpolarization activated cyclic nucleotide-gated... INVS 0.034 inversin isoform a INVS 0.034 inversin isoform b LOC100292378 0.031 PREDICTED: hypothetical protein LTBP4 0.031 latent transforming growth factor beta binding prot... LTBP4 0.031 latent transforming growth factor beta binding prot... LTBP4 0.031 latent transforming growth factor beta binding prot... LOC65998 0.031 hypothetical protein LOC65998 LOC645722 0.031 PREDICTED: similar to hCG2037003 COL3A1 0.028 collagen type III alpha 1 preproprotein TMEM132E 0.028 transmembrane protein 132E ADAMTS14 0.028 ADAM metallopeptidase with thrombospondin type 1 mot... ADAMTS14 0.028 ADAM metallopeptidase with thrombospondin type 1 mo... SSH1 0.028 slingshot 1 isoform 1 GPR135 0.028 G protein-coupled receptor 135 SSPO 0.028 SCO-spondin NONO 0.028 non-POU domain containing, octamer-binding isoform ... NONO 0.028 non-POU domain containing, octamer-binding isoform ... NONO 0.028 non-POU domain containing, octamer-binding isoform ... NONO 0.028 non-POU domain containing, octamer-binding isoform 1... RLTPR 0.024 RGD motif, leucine rich repeats, tropomodulin domain... CXXC1 0.024 CXXC finger 1 (PHD domain) isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
